Meglutol
- MEGLUTOL
- 503-49-1
- 3-Hydroxy-3-methylglutaric acid
- 3-Hydroxy-3-methylpentanedioic acid
- Dicrotalic acid
- Create:2004-09-16
- Modify:2025-01-25
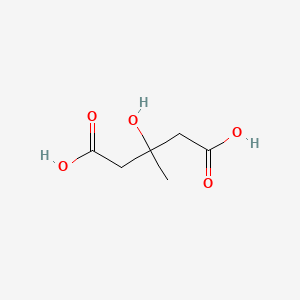
- 3 Hydroxy 3 methylglutaric Acid
- 3 Hydroxy 3 methylpentanedioic Acid
- 3-Hydroxy-3-methylglutaric Acid
- 3-Hydroxy-3-methylpentanedioic Acid
- Acid, 3-Hydroxy-3-methylglutaric
- Acid, 3-Hydroxy-3-methylpentanedioic
- beta Hydroxy beta Methylglutarate
- beta-Hydroxy-beta-Methylglutarate
- Meglutol
- MEGLUTOL
- 503-49-1
- 3-Hydroxy-3-methylglutaric acid
- 3-Hydroxy-3-methylpentanedioic acid
- Dicrotalic acid
- Pentanedioic acid, 3-hydroxy-3-methyl-
- Lipoglutaren
- Medroglutaric acid
- HMGA
- beta-Hydroxy-beta-methylglutaric acid
- 3-hydroxy-3-methyl-glutaric acid
- CB 337
- Meglutolum
- Meglutol [USAN:INN]
- CB-337
- Meglutolum [INN-Latin]
- 3-Hydroxy-3-methylglutarate
- Glutaric acid, 3-hydroxy-3-methyl-
- 3-Hydorxy-3-methylglutaric acid
- 3-Hydroxymethylglutarate
- NSC-361411
- CLA99KCD53
- CHEMBL50444
- 3-Hydroxy-3-methylpentane-1,5-dioic Acid
- HMG
- CHEBI:16831
- (S)-Meglutol
- Dicrotalic acid;3-Hydroxy-3-methylglutaric acid
- (S)-3-Hydroxy-3-methylglutaric acid
- EINECS 207-971-1
- MFCD00002712
- UNII-CLA99KCD53
- NSC 361411
- BRN 1769194
- Dicrotalate
- Menotrophin?
- beta-Hydroxy-beta-Methylglutarate
- MEGLUTOL [USAN]
- Meglutol (USAN/INN)
- MEGLUTOL [INN]
- MEGLUTOL [MI]
- MEGLUTOL [MART.]
- bmse000335
- b-Hydroxy-b-methylglutarate
- 3-Methyl-3-hydroxyglutarate
- SCHEMBL28443
- 4-03-00-01166 (Beilstein Handbook Reference)
- MLS001066390
- b-Hydroxy-b-methylglutaric acid
- GTPL2960
- MEGxp0_001907
- 3-Methyl-3-hydroxyglutaric acid
- ACon1_000502
- DTXSID90198304
- HMS3264B09
- Pharmakon1600-01506183
- HY-B1189
- 3-HYDROXY-3-METHYLPENTANE-1
- BDBM50160720
- NSC361411
- NSC760412
- AKOS000280741
- CCG-213633
- CS-4798
- DB04377
- NSC-760412
- NCGC00169018-01
- AS-63885
- GLUTARIC ACID, BETA-HYDROXY METHYL
- SMR000471861
- 3-Hydroxy-3-methylglutaric acid, >=95%
- DB-071173
- H0436
- NS00014740
- (3R)-3,5,5-trihydroxy-3-methylpentanoic acid
- C03761
- D04897
- F17049
- AB00698060_04
- Dicrotalic acid; 3-Hydroxy-3-methylglutaric acid
- Q524670
- SR-01000763754
- 3-Hydroxy-3-methylpentane-1 pound not5-dioic Acid
- SR-01000763754-2
- A90FC159-16ED-49CA-9A7F-D90B0A5B3EBC
- BRD-K00495673-001-05-2
- 3-Hydroxy-3-methylglutaric acid, purum, >=95.0% (GC)
- XII
135.9 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
126.5 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
126.9 Ų [M-H]-
135.9 Ų [M+Na]+
147.1 100
247.1 74.49
199.05 49.03
231.0 45.04
115.05 41.30
147.0 100
115.0 57.16
109.0 45.15
247.0 37.74
199.0 35.74
57.03433 100
59.01358 29.70
99.045 13.50
55.05464 9.20
41.00114 1.60
57.03942 100
99.05109 82.60
101.031 50.90
161.05101 35.20
59.01885 27.40
161.1 999
99 91
125 60
100.8 51
57.5 3
99 999
101.2 443
57.3 352
161.3 147
81.2 130
147.1 999
247.1 744
199.05 490
231 450
115.05 413

H315 (100%): Causes skin irritation [Warning Skin corrosion/irritation]
H319 (100%): Causes serious eye irritation [Warning Serious eye damage/eye irritation]
P264, P264+P265, P280, P302+P352, P305+P351+P338, P321, P332+P317, P337+P317, and P362+P364
(The corresponding statement to each P-code can be found at the GHS Classification page.)
Skin Irrit. 2 (100%)
Eye Irrit. 2 (100%)
Silke Matysik, Caroline Ivanne Le Roy, Gerhard Liebisch, Sandrine Paule Claus. Metabolomics of fecal samples: A practical consideration. Trends in Food Science & Technology. Vol. 57, Part B, Nov. 2016, p.244-255: http://www.sciencedirect.com/science/article/pii/S0924224416301984
PubMed: 6157502, 11129331, 28583327, 15505778, 25557019, 23705938, 1886403, 12072887, 19893767
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=NPOAOTPXWNWTSH-UHFFFAOYSA-N
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/3-Hydroxy-3-methylglutaric acidhttps://commonchemistry.cas.org/detail?cas_rn=503-49-1
- ChemIDplusMeglutol [USAN:INN]https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0000503491ChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_use
- DTP/NCILICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuse
- EPA DSSToxCompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-noticeMeglutol (EC: 207-971-1)https://echa.europa.eu/information-on-chemicals/cl-inventory-database/-/discli/details/46857
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linking
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citing3-Hydroxymethylglutaric acidhttp://www.hmdb.ca/metabolites/HMDB0000355HMDB0000355_cms_30141https://hmdb.ca/metabolites/HMDB0000355#spectra
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/Meglutol | 3-hydroxy-3-methylglutaric acidNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBI3-hydroxy-3-methylglutaric acidhttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:16831
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/LOTUS Treehttps://lotus.naturalproducts.net/
- Open TargetsLICENSEDatasets generated by the Open Targets Platform are freely available for download.https://platform-docs.opentargets.org/licence
- Toxin and Toxin Target Database (T3DB)LICENSET3DB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (T3DB) and the original publication.http://www.t3db.ca/downloads3-Hydroxymethylglutaric acidhttp://www.t3db.ca/toxins/T3D4262
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- ClinicalTrials.govLICENSEThe ClinicalTrials.gov data carry an international copyright outside the United States and its Territories or Possessions. Some ClinicalTrials.gov data may be subject to the copyright of third parties; you should consult these entities for any additional terms of use.https://clinicaltrials.gov/ct2/about-site/terms-conditions#Use
- Therapeutic Target Database (TTD)3-Hydroxy-3-Methyl-Glutaric Acidhttps://idrblab.net/ttd/data/drug/details/D0IJ7H
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutbeta-Hydroxy-beta-methylglutaric acidhttps://foodb.ca/compounds/FDB004207
- MassBank Europe3-Hydroxy-3-Methylglutaratehttps://massbank.eu/MassBank/Result.jsp?inchikey=NPOAOTPXWNWTSH-UHFFFAOYSA-N
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- SpectraBase3-HYDROXY-3-METHYLGLUTARIC ACIDhttps://spectrabase.com/spectrum/FbVh5WRAQan3-Hydroxy-3-methylglutaric acidhttps://spectrabase.com/spectrum/7Knj2PHcVhebeta-Hydroxy-beta-methylglutaric acidhttps://spectrabase.com/spectrum/8OKQxMSAabW3-Hydroxy-3-methylglutaric acidhttps://spectrabase.com/spectrum/BhJt7exjhD03-HYDROXY-3-METHYLGLUTARIC ACIDhttps://spectrabase.com/spectrum/JSmtE7dJstM3-Hydroxy-3-methylglutaric acidhttps://spectrabase.com/spectrum/3AE4jhNCmsu3-HYDROXY-3-METHYLGLUTARIC ACIDhttps://spectrabase.com/spectrum/FOPToxAuOtS3-Hydroxy-3-methylglutaric acidhttps://spectrabase.com/spectrum/FUREvy0zCmt3-Hydroxy-3-methylglutaric acidhttps://spectrabase.com/spectrum/5EpSWkuhk87
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-law3-Hydroxy-3-methylglutaric acidhttp://www.nist.gov/srd/nist1a.cfm
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlAnatomical Therapeutic Chemical (ATC) classificationhttp://www.genome.jp/kegg-bin/get_htext?br08303.keg
- KNApSAcK Species-Metabolite Database3-Hydroxy-3-methylglutaric acidhttp://www.knapsackfamily.com/knapsack_core/info.php?sname=C_ID&word=C00001187
- Natural Product Activity and Species Source (NPASS)
- MarkerDBLICENSEThis work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.https://markerdb.ca/3-Hydroxymethylglutaric acidhttps://markerdb.ca/chemicals/215
- Metabolomics Workbench3-Hydroxy-3-methylglutaric acidhttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=37215
- NCI Thesaurus (NCIt)LICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuseNCI Thesaurushttps://ncit.nci.nih.gov
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Springer Nature
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- WHO Anatomical Therapeutic Chemical (ATC) ClassificationLICENSEUse of all or parts of the material requires reference to the WHO Collaborating Centre for Drug Statistics Methodology. Copying and distribution for commercial purposes is not allowed. Changing or manipulating the material is not allowed.https://www.whocc.no/copyright_disclaimer/
- Wikidata
- Wikipedia
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmlHypolipidemic Agentshttps://www.ncbi.nlm.nih.gov/mesh/68000960Anticholesteremic Agentshttps://www.ncbi.nlm.nih.gov/mesh/68000924Enzyme Inhibitorshttps://www.ncbi.nlm.nih.gov/mesh/68004791Hydroxymethylglutaryl-CoA Reductase Inhibitorshttps://www.ncbi.nlm.nih.gov/mesh/68019161
- PubChem
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 388497179https://pubchem.ncbi.nlm.nih.gov/substance/388497179











