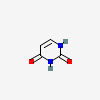
Uracil
- uracil
- 66-22-8
- 2,4-Dihydroxypyrimidine
- 2,4(1H,3H)-Pyrimidinedione
- pyrimidine-2,4(1H,3H)-dione
- Create:2004-09-16
- Modify:2024-12-27
 Pyrimidine (subclass of).
Pyrimidine (subclass of).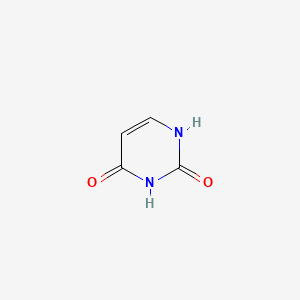
- uracil
- 66-22-8
- 2,4-Dihydroxypyrimidine
- 2,4(1H,3H)-Pyrimidinedione
- pyrimidine-2,4(1H,3H)-dione
- pyrimidine-2,4-diol
- Pyrod
- 2,4-Pyrimidinediol
- 2,4-Dioxopyrimidine
- Hybar X
- 2,4-Pyrimidinedione
- Pirod
- 51953-14-1
- 1H-Pyrimidine-2,4-dione
- RU 12709
- 1,2,3,4-tetrahydropyrimidine-2,4-dione
- Uracil [USAN]
- Urazil
- Ura
- MFCD00006016
- CCRIS 3077
- CHEBI:17568
- NSC 3970
- NSC-3970
- 2,4-Pyrimidinediol (9CI)
- AI3-25470
- Uracyl
- 2-Hydroxy-4(3H)-pyrimidinone
- SQ 6201
- SQ 7726
- SQ 8493
- SQ-6201
- SQ-7726
- SQ-8493
- BMS 205603-01
- BMS-205603-01
- UNII-56HH86ZVCT
- EINECS 200-621-9
- 56HH86ZVCT
- 24897-51-6
- Lamivudine impurity e
- 2-Hydroxy-4(1H)-pyrimidinone
- 66255-05-8
- DTXSID4021424
- NSC3970
- Lamivudine impurity e rs
- DTXCID101424
- 144104-68-7
- 4-Hydroxy-2(1H)-pyrimidinone
- Fluorouracil specified compound c
- NCGC00181030-01
- URACIL (USP-RS)
- URACIL [USP-RS]
- URACIL (MART.)
- URACIL [MART.]
- 4-Hydroxyuracil
- CAS-66-22-8
- CID 5274267
- LAMIVUDINE IMPURITY F (EP IMPURITY)
- LAMIVUDINE IMPURITY F [EP IMPURITY]
- FLUOROURACIL IMPURITY C (EP IMPURITY)
- FLUOROURACIL IMPURITY C [EP IMPURITY]
- LAMIVUDINE IMPURITY E (USP IMPURITY)
- LAMIVUDINE IMPURITY E [USP IMPURITY]
- Uracil (8CI)
- Uracil [USAN:JAN]
- 4(3H)-Pyrimidinone, 2-hydroxy-
- FLUOROURACIL SPECIFIED COMPOUND C (USP IMPURITY)
- FLUOROURACIL SPECIFIED COMPOUND C [USP IMPURITY]
- 8h-uracil
- hydroxypyrimidinone
- 2(1H)-Pyrimidinone, 4-hydroxy-
- 2(1H)-Pyrimidinone, 6-hydroxy-
- Uracil (Standard)
- 2,6-Dioxypyrimidin
- Uracil,(S)
- 2,4-Dioxypyrimidine
- 2,4(1H,3H)-Pyrimidinedione (9CI)
- 66224-60-0
- pyrimidine-2,4-dione
- 2, 4-Dioxopyrimidine
- Fluorouracil Impurity C
- Uracil, 99%
- 1ui0
- Uracil (JAN/USAN)
- 2,3H)-Pyrimidinedione
- 2,6-Dihydroxypyrimidine
- URACIL [JAN]
- URACIL [WHO-DD]
- URACIL [MI]
- bmse000187
- bmse000940
- CHEMBL566
- Epitope ID:120356
- NCIMech_000782
- SCHEMBL8235
- Uracil, >=99.0%
- pyrimidine, 2,4-dihydroxy-
- MLS001304993
- GTPL4560
- 2,4-(1h,3h)-pyrimidinedione
- 2-hydroxy-4-(1H)-pyrimidione
- 2-hydroxy-4-(3H)-pyrimidione
- 4-hydroxy-2-(1H)-pyrimidione
- HY-I0960R
- Uracil, >=99.0% (T)
- DTXSID501007260
- HMS2234E19
- HMS3264C13
- HMS3373E18
- HMS3652N05
- Pharmakon1600-01502345
- (4-Hydroxypyrimidin-2-yl)oxidanyl
- 4(1H)-Pyrimidinone, 2-hydroxy-
- BCP26546
- HY-I0960
- NSC29742
- Tox21_112680
- Tox21_201023
- BDBM50549809
- CCG-35866
- NSC-29742
- NSC759649
- s4177
- STK301734
- STL124066
- AKOS000119989
- AKOS002303991
- Tox21_112680_1
- 4(3H)-Pyrimidinone,2-hydroxy-(9ci)
- CCG-213042
- CS-W020104
- DB03419
- NSC-759649
- PS-5279
- SB55489
- SB55884
- NCGC00181030-02
- NCGC00247663-01
- NCGC00258576-01
- Uracil, Vetec(TM) reagent grade, 98%
- 51953-19-6
- NCI60_003718
- Pyrimidine-2,4(1H,3H)-dione (Uracil)
- SMR000752912
- SY008943
- DB-030518
- DB-103964
- DB-268940
- DB-268966
- DB-272084
- DB-272202
- NS00002070
- SW220239-1
- U0013
- EN300-17138
- Uracil, suitable for cell culture, BioReagent
- C00106
- D00027
- SBI-0053640.0002
- AB00171810_03
- AB00171810_04
- AB00918623-05
- AC-907/30002021
- Q182990
- BRD-K80129304-001-08-1
- BRD-K80129304-001-09-9
- Z56889474
- F1796-0008
- E2FC11E5-1887-46DF-B415-82313CE9B2BD
- Uracil, United States Pharmacopeia (USP) Reference Standard
- Fluorouracil impurity C, European Pharmacopoeia (EP) Reference Standard
- InChI=1/C4H4N2O2/c7-3-1-2-5-4(8)6-3/h1-2H,(H2,5,6,7,8
- Uracil, Pharmaceutical Secondary Standard; Certified Reference Material
113.5 Ų [M-H]-
126.2 Ų [M+H]+
- 15N nuclear magnetic resonance spectrum
- Coriolis coupling
- Schoenflies notation
- Boiling point
- Centrifugal distortion
- Chemical bond
- Chemical shift
- Crystal structure
- Density
- Diamagnetic susceptibility
- Equilibrium structure
- Formula unit
- Formula weight
- Heat of sublimation
- Internuclear distance
- Lineshape
- Magnetic susceptibility
- Molecular structure
- Nuclear quadrupole coupling
- Nuclear quadrupole resonance spectroscopy
- Point group
- Quadrupole coupling
- Rotation-vibration spectrum
- Rotational excitation cross section
- Space group
- Spin-spin coupling constant
- Unit cell
- Unit cell parameter
- Vapor pressure
99.0 1
241.0 0.81
255.0 0.45
256.0 0.35
113.0 0.25
112.0 99.99
69.0 69.75
42.0 58.56
17.0 40.90
28.0 37.37
41.99852 100
111.0234 58.81
74.96782 1.70
41.99852 100
111.0197 2.11
68.01588 1.73
111.2 999
67 13
42 4
111 999
41.8 467
67 221
46.2 8
112 999
69 698
42 586
17 409
28 374
112 999
69 472
42 395
28 368
41 249
 Pyrimidine (subclass of)
Pyrimidine (subclass of)- All Tissues
- Placenta
- Prostate
<b>Use (kg; approx.) in Germany (2009):</b> >10
<b>Consumption (g per capita; approx.) in Germany (2009):</b> 0.000122
<b>Calculated removal (%):</b> 75.1

H315 (10.5%): Causes skin irritation [Warning Skin corrosion/irritation]
H319 (10.5%): Causes serious eye irritation [Warning Serious eye damage/eye irritation]
H335 (10.5%): May cause respiratory irritation [Warning Specific target organ toxicity, single exposure; Respiratory tract irritation]
P261, P264, P264+P265, P271, P280, P302+P352, P304+P340, P305+P351+P338, P319, P321, P332+P317, P337+P317, P362+P364, P403+P233, P405, and P501
(The corresponding statement to each P-code can be found at the GHS Classification page.)
Aggregated GHS information provided per 19 reports by companies from 5 notifications to the ECHA C&L Inventory. Each notification may be associated with multiple companies.
Reported as not meeting GHS hazard criteria per 16 of 19 reports by companies. For more detailed information, please visit ECHA C&L website.
There are 3 notifications provided by 3 of 19 reports by companies with hazard statement code(s).
Information may vary between notifications depending on impurities, additives, and other factors. The percentage value in parenthesis indicates the notified classification ratio from companies that provide hazard codes. Only hazard codes with percentage values above 10% are shown.
Skin Irrit. 2 (10.5%)
Eye Irrit. 2A (10.5%)
STOT SE 3 (10.5%)
IMAP assessments - 2,4(1H,3H)-Pyrimidinedione: Environment tier I assessment
IMAP assessments - 2,4(1H,3H)-Pyrimidinedione: Human health tier I assessment
Merck Manual of Diagnosis and Therapy.
David F. Putnam Composition and Concentrative Properties of Human Urine. NASA Contractor Report. July 1971
Geigy Scientific Tables, 8th Rev edition, pp. 130. Edited by C. Lentner, West Cadwell, N.J.: Medical education Div., Ciba-Geigy Corp. Basel, Switzerland c1981-1992.
Geigy Scientific Tables, 8th Rev edition, pp. 165-177. Edited by C. Lentner, West Cadwell, N.J.: Medical education Div., Ciba-Geigy Corp., Basel, Switzerland c1981-1992.
National Health and Nutrition Examination Survey (NHANES Survey) 2013
Geigy Scientific Tables, 8th Rev edition, pp. 80-82. Edited by C. Lentner, West Cadwell, N.J.: Medical education Div., Ciba-Geigy Corp., Basel, Switzerland c1981-1992.
PubMed: 624188, 6796772, 2311630, 26123990, 28659245
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
PubMed: 7680187, 5471650
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
Silke Matysik, Caroline Ivanne Le Roy, Gerhard Liebisch, Sandrine Paule Claus. Metabolomics of fecal samples: A practical consideration. Trends in Food Science & Technology. Vol. 57, Part B, Nov. 2016, p.244-255: http://www.sciencedirect.com/science/article/pii/S0924224416301984
PubMed: 14705962, 8581129, 2109146
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=ISAKRJDGNUQOIC-UHFFFAOYSA-N
- Australian Industrial Chemicals Introduction Scheme (AICIS)2,4(1H,3H)-Pyrimidinedionehttps://services.industrialchemicals.gov.au/search-assessments/
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/2,4-Pyrimidinediolhttps://commonchemistry.cas.org/detail?cas_rn=51953-14-14(1H)-Pyrimidinone, 2-hydroxy-https://commonchemistry.cas.org/detail?cas_rn=51953-20-92,4(1H,3H)-Pyrimidinedione-14Chttps://commonchemistry.cas.org/detail?cas_rn=2920-92-52,4(1H,3H)-Pyrimidinedione, dimerhttps://commonchemistry.cas.org/detail?cas_rn=28806-15-7
- ChemIDplusChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_use
- DTP/NCILICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuse
- EPA Chemicals under the TSCA2,4(1H,3H)-Pyrimidinedionehttps://www.epa.gov/chemicals-under-tscaEPA TSCA Classificationhttps://www.epa.gov/tsca-inventory
- EPA DSSTox(4-Hydroxypyrimidin-2-yl)oxidanylhttps://comptox.epa.gov/dashboard/DTXSID501007260CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-notice
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linking
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingHMDB0000300_cms_1056https://hmdb.ca/metabolites/HMDB0000300#spectra
- New Zealand Environmental Protection Authority (EPA)LICENSEThis work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International licence.https://www.epa.govt.nz/about-this-site/general-copyright-statement/
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/URACILNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBI
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/LOTUS Treehttps://lotus.naturalproducts.net/
- Open TargetsLICENSEDatasets generated by the Open Targets Platform are freely available for download.https://platform-docs.opentargets.org/licence
- Yeast Metabolome Database (YMDB)
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- ClinicalTrials.govLICENSEThe ClinicalTrials.gov data carry an international copyright outside the United States and its Territories or Possessions. Some ClinicalTrials.gov data may be subject to the copyright of third parties; you should consult these entities for any additional terms of use.https://clinicaltrials.gov/ct2/about-site/terms-conditions#Use
- Comparative Toxicogenomics Database (CTD)LICENSEIt is to be used only for research and educational purposes. Any reproduction or use for commercial purpose is prohibited without the prior express written permission of NC State University.http://ctdbase.org/about/legal.jsp
- Therapeutic Target Database (TTD)
- Crystallography Open Database (COD)LICENSEAll data in the COD and the database itself are dedicated to the public domain and licensed under the CC0 License. Users of the data should acknowledge the original authors of the structural data.https://creativecommons.org/publicdomain/zero/1.0/
- IUPAC Digitized pKa DatasetPyrimidine-2,4(1H,3H)-dionehttps://github.com/IUPAC/Dissociation-Constants
- ECI Group, LCSB, University of Luxembourguracil
- KNApSAcK Species-Metabolite Database
- Natural Product Activity and Species Source (NPASS)
- West Coast Metabolomics Center-UC DavisUracil
- EPA Chemical and Products Database (CPDat)EPA CPDat Classificationhttps://www.epa.gov/chemical-research/chemical-and-products-database-cpdat
- Haz-Map, Information on Hazardous Chemicals and Occupational DiseasesLICENSECopyright (c) 2022 Haz-Map(R). All rights reserved. Unless otherwise indicated, all materials from Haz-Map are copyrighted by Haz-Map(R). No part of these materials, either text or image may be used for any purpose other than for personal use. Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited without prior written permission.https://haz-map.com/About
- EU Clinical Trials Register
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/about
- MassBank Europe
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-law
- SpectraBasePYRIMIDINE-2,4-DIOLhttps://spectrabase.com/spectrum/Jkx0j9ypTy12,4(1H,3H)-PYRIMIDINEDIONEhttps://spectrabase.com/spectrum/4eQ1eX79fYLPyrimidine-2,4-diolhttps://spectrabase.com/spectrum/A5YAYrd0tXVPyrimidine-2,4-diolhttps://spectrabase.com/spectrum/8HhmoxOGMfRISAKRJDGNUQOIC-UHFFFAOYSA-Nhttps://spectrabase.com/spectrum/Lw6BeD22B93
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlCompounds with biological roleshttp://www.genome.jp/kegg-bin/get_htext?br08001.keg
- Kruve Lab, Ionization & Mass Spectrometry, Stockholm Universityuracil
- MarkerDBLICENSEThis work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.https://markerdb.ca/
- Metabolomics Workbench
- NCI Thesaurus (NCIt)LICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuseNCI Thesaurushttps://ncit.nci.nih.gov
- NIPH Clinical Trials Search of Japan
- NMRShiftDB
- PharmGKBLICENSEPharmGKB data are subject to the Creative Commons Attribution-ShareALike 4.0 license (https://creativecommons.org/licenses/by-sa/4.0/).https://www.pharmgkb.org/page/policies
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- Springer Nature
- SpringerMaterials2,4(1H,3H)-Pyrimidinedionehttps://materials.springer.com/substanceprofile/docs/smsid_maesnpcdqbmhurvw
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- Wikidata
- Wikipedia
- Wiley
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.html
- PubChem
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- EPA Substance Registry ServicesEPA SRS List Classificationhttps://sor.epa.gov/sor_internet/registry/substreg/LandingPage.do
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403382681https://pubchem.ncbi.nlm.nih.gov/substance/403382681
- NCBI