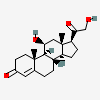
Corticosterone
- CORTICOSTERONE
- 50-22-6
- 17-Deoxycortisol
- Reichstein's substance H
- Kendall's compound B
- Create:2004-09-16
- Modify:2025-01-18
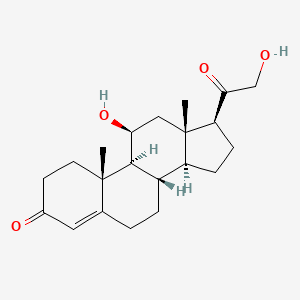
- CORTICOSTERONE
- 50-22-6
- 17-Deoxycortisol
- Reichstein's substance H
- Kendall's compound B
- Corticosteron
- 11beta,21-Dihydroxyprogesterone
- Reichstein's B
- 11-Hydroxycorticoaldosterone
- (11beta)-11,21-Dihydroxypregn-4-ene-3,20-dione
- 11beta,21-Dihydroxy-4-pregnene-3,20-dione
- 4-Pregnene-11beta,21-diol-3,20-dione
- 11,21-Dihydroxyprogesterone
- 11Beta,21-dihydroxypregn-4-ene-3,20-dione
- NSC-9705
- 11-beta,21-Dihydroxypregn-3,20-dione
- CHEBI:16827
- NSC9705
- 4-Pregnene-11-beta,21-diol-3,20-dione
- Pregn-4-ene-3,20-dione, 11,21-dihydroxy-, (11beta)-
- Pregn-4-ene-3,20-dione, 11beta,21-dihydroxy-
- Pregn-4-ene-3,20-dione, 11-beta,21-dihydroxy-
- (11-beta)-11,21-Dihydroxypregn-4-ene-3,20-dione
- MLS000028536
- CHEMBL110739
- (8S,9S,10R,11S,13S,14S,17S)-11-hydroxy-17-(2-hydroxyacetyl)-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta[a]phenanthren-3-one
- DTXSID6022474
- W980KJ009P
- 11.beta.,21-Dihydroxyprogesterone
- NCGC00022472-05
- SMR000058318
- 4-Pregnene-11.beta.,21-diol-3,20-dione
- Corticosterone 100 microg/mL in Acetonitrile
- DTXCID702474
- (11.beta.)-11,21-Dihydroxypregn-4-ene-3,20-dione
- (8S,9S,10R,11S,13S,14S,17S)-11-hydroxy-17-(2-hydroxyacetyl)-10,13-dimethyl-1,2,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-3H-cyclopenta[a]phenanthren-3-one
- 11,12-Dihydroxyprogesterone
- CCRIS 6753
- 11-beta,21-Dihydroxyprogesterone
- Compound B nach Kendall
- SR-01000075748
- NSC 9705
- EINECS 200-019-6
- Substanz H nach Reichstein
- BRN 2339601
- Cortico
- UNII-W980KJ009P
- Compd B
- CAS-50-22-6
- Prestwick_672
- Kendalls Compound B
- MFCD00037715
- Pregn-4-ene-3,20-dione, 11,21-dihydroxy-, (11b)-
- Reichsteins Substance H
- 21-dihydroxyprogesterone
- (11-BETA)-11,21-DIHYDROXY-PREGN-4-ENE-3,20-DIONE
- Opera_ID_1519
- Prestwick0_000432
- Prestwick1_000432
- Prestwick2_000432
- Prestwick3_000432
- Corticosterone (Standard)
- Corticosterone, >=92%
- 11-.beta.,20-dione
- bmse000669
- Epitope ID:152207
- CORTICOSTERONE [MI]
- BIDD:PXR0063
- Lopac0_000220
- SCHEMBL22612
- BSPBio_000444
- 4-Pregnene-11 Corticosteron
- MLS001074095
- MLS001424305
- Pregn-4-ene-3,20-dione, 11,21-dihydroxy-, (11.beta.)-
- BIDD:ER0495
- SPBio_002383
- BPBio1_000490
- GTPL2869
- HY-B1618R
- Corticosterone, BRN 2339601
- 4-Pregnene-11.beta.,20-dione
- HMS1569G06
- HMS2052G21
- HMS2090A08
- HMS2096G06
- HMS2231F16
- HMS3260L21
- HMS3713G06
- HMS3885H22
- Corticosterone, 1mg/ml in Methanol
- HY-B1618
- Tox21_110879
- Tox21_201366
- Tox21_303642
- Tox21_500220
- BDBM50170653
- CMC_13412
- HB2808
- HSCI1_000383
- LMST02030186
- s4752
- STL564760
- Corticosterone, >=98.5% (HPLC)
- AKOS016008541
- Tox21_110879_1
- 11-b,21-Dihydroxypregn-3,20-dione
- CCG-101146
- CS-5105
- DB04652
- LP00220
- NC00396
- SDCCGSBI-0050208.P002
- SMP1_000079
- NCGC00022472-06
- NCGC00022472-07
- NCGC00022472-08
- NCGC00022472-10
- NCGC00022472-17
- NCGC00091029-01
- NCGC00256539-01
- NCGC00258918-01
- NCGC00260905-01
- 11|A
- AS-75743
- DA-52062
- 11,21-Dihydroxypregn-4-ene-3,20-dione
- 11-.beta.,21-Dihydroxypregn-3,20-dione
- 11b,21-Dihydroxy-4-pregnene-3,20-dione
- Pregn-4-ene-3, 11.beta.,21-dihydroxy-
- 4-Pregnen-11.beta.,21-diol-3,20-dione
- EU-0100220
- NS00000569
- C 2505
- C02140
- D94656
- Preg-4-ene-3,20-dione,11-b,21-dihydroxy-
- 11.beta.,21-dihydroxypregn-4-ene-3,20-dione
- Corticosterone, VETRANAL(TM), analytical standard
- Q422543
- SR-01000000082
- Pregn-4-ene-3,20-dione, 11.beta.,21-dihydroxy-
- SR-01000000082-3
- SR-01000075748-1
- SR-01000075748-4
- W-105977
- WLN: L E5 B666 OV MUTJ A1 CQ E1 FV1Q
- (11beta)-11,21-Dihydroxypregn-4-ene-3,2 0-dione
- BRD-K73589401-001-04-6
- BRD-K73589401-001-23-6
- Pregn-4-ene-3, 11,21-dihydroxy-, (11.beta.)-
- (9beta,11alpha)-11,21-dihydroxypregn-4-ene-3,20-dione
- 11,21-Dihydroxypregn-4-ene-3,20-dione, (11.beta.)- #
- 4793EB71-D789-498E-B641-C5C1C377B9FF
- 17-Deoxycortisol;11,21-Dihydroxyprogesterone;Kendall's compound B
- Pregn-4-ene-3,20-dione, 11,21-dihydroxy-, (11-beta)- (9CI)
- Kendall's Compound B; 4-Pregnene-11beta,21-diol-3,20-dione; Reichstein's Substance H
186.8 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
211.8 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
208.4 Ų [M+K]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
186.9 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
186.8 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with Agilent tune mix (Agilent)]
186.46 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with Agilent tune mix (Agilent)]
209 Ų [M+K]+
212.2 Ų [M+Na]+
187 Ų [M+H]+
186.8 Ų [M-H]-
315.0 99.99
269.0 71.05
316.0 22.55
227.0 17.17
270.0 15.53
89.0 1
103.0 0.78
91.0 0.59
79.0 0.37
105.0 0.35
347.3 100
329.201 6.44
311.15 2
121.033 1.95
293.115 1.64
121.009 100
105.067 80.62
91.093 74.42
97.081 72.87
93.084 65.12
121.0642 999
97.0642 409
123.0792 341
119.0853 316
147.08 314
347.2215 999
329.2111 485
293.1898 170
311.2003 161
121.065 151
315 999
269 711
316 226
227 172
270 155
121.065620 100
347.220886 80.27
329.210632 67.50
91.056419 46.44
105.071358 45.37
- Adipose Tissue
- Adrenal Cortex
- Adrenal Gland
- Brain
- Epidermis
- Fibroblasts
- Intestine
- Kidney
- Liver
- Neuron
- Ovary
- Placenta
- Platelet
- Spleen
- Testis
- Cytoplasm
- Endoplasmic reticulum
- Extracellular
- Membrane
- Mitochondria
- 11-beta-hydroxylase deficiency (CYP11B1)
- 17-alpha-hydroxylase deficiency (CYP17)
- 21-hydroxylase deficiency (CYP21)
- 3-Beta-Hydroxysteroid Dehydrogenase Deficiency
- Adrenal Hyperplasia Type 3 or Congenital Adrenal Hyperplasia due to 21-hydroxylase Deficiency
- Adrenal Hyperplasia Type 5 or Congenital Adrenal Hyperplasia due to 17 Alpha-hydroxylase Deficiency
- Apparent mineralocorticoid excess syndrome
- Congenital Lipoid Adrenal Hyperplasia (CLAH) or Lipoid CAH
- Corticosterone methyl oxidase I deficiency (CMO I)
- Corticosterone methyl oxidase II deficiency - CMO II
- Total 11 pathways, visit the HMDB page for details

P261, P272, P280, P302+P352, P321, P333+P317, P362+P364, and P501
(The corresponding statement to each P-code can be found at the GHS Classification page.)
Aggregated GHS information provided per 40 reports by companies from 2 notifications to the ECHA C&L Inventory.
Information may vary between notifications depending on impurities, additives, and other factors. The percentage value in parenthesis indicates the notified classification ratio from companies that provide hazard codes. Only hazard codes with percentage values above 10% are shown.
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=OMFXVFTZEKFJBZ-HJTSIMOOSA-N
- BindingDBLICENSEAll data curated by BindingDB staff are provided under the Creative Commons Attribution 3.0 License (https://creativecommons.org/licenses/by/3.0/us/).https://www.bindingdb.org/rwd/bind/info.jsp(11beta)-11,21-dihydroxypregn-4-ene-3,20-dionehttps://www.bindingdb.org/rwd/bind/chemsearch/marvin/MolStructure.jsp?monomerid=50170653
- Comparative Toxicogenomics Database (CTD)LICENSEIt is to be used only for research and educational purposes. Any reproduction or use for commercial purpose is prohibited without the prior express written permission of NC State University.http://ctdbase.org/about/legal.jspCorticosteronehttps://ctdbase.org/detail.go?type=chem&acc=D003345
- Drug Gene Interaction database (DGIdb)LICENSEThe data used in DGIdb is all open access and where possible made available as raw data dumps in the downloads section.http://www.dgidb.org/downloadsCORTICOSTERONEhttps://www.dgidb.org/drugs/drugbank:DB04652IRPAGRATINIBhttps://www.dgidb.org/drugs/iuphar.ligand:12869
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_useCorticosteronehttps://www.drugbank.ca/drugs/DB04652
- IUPHAR/BPS Guide to PHARMACOLOGYLICENSEThe Guide to PHARMACOLOGY database is licensed under the Open Data Commons Open Database License (ODbL) https://opendatacommons.org/licenses/odbl/. Its contents are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (http://creativecommons.org/licenses/by-sa/4.0/)https://www.guidetopharmacology.org/about.jsp#licenseGuide to Pharmacology Target Classificationhttps://www.guidetopharmacology.org/targets.jsp
- Therapeutic Target Database (TTD)(11-BETA)-11,21-DIHYDROXY-PREGN-4-ENE-3,20-DIONEhttps://idrblab.net/ttd/data/drug/details/D03TNT
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/Corticosteronehttps://commonchemistry.cas.org/detail?cas_rn=50-22-6
- ChemIDplusChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- DTP/NCILICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuse
- EPA DSSToxCorticosteronehttps://comptox.epa.gov/dashboard/DTXSID6022474CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-noticeCorticosterone (EC: 200-019-6)https://echa.europa.eu/information-on-chemicals/cl-inventory-database/-/discli/details/42584
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linking
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingCorticosteronehttp://www.hmdb.ca/metabolites/HMDB0001547HMDB0001547_cms_29606https://hmdb.ca/metabolites/HMDB0001547#spectra
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/CorticosteroneNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBI
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/Corticosteronehttps://www.wikidata.org/wiki/Q422543LOTUS Treehttps://lotus.naturalproducts.net/
- NCI Thesaurus (NCIt)LICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuseNCI Thesaurushttps://ncit.nci.nih.gov
- Open TargetsLICENSEDatasets generated by the Open Targets Platform are freely available for download.https://platform-docs.opentargets.org/licenceCORTICOSTERONEhttps://platform.opentargets.org/drug/CHEMBL110739
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- ClinicalTrials.govLICENSEThe ClinicalTrials.gov data carry an international copyright outside the United States and its Territories or Possessions. Some ClinicalTrials.gov data may be subject to the copyright of third parties; you should consult these entities for any additional terms of use.https://clinicaltrials.gov/ct2/about-site/terms-conditions#Use
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutCorticosteronehttps://foodb.ca/compounds/FDB022684
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-lawCorticosteronehttp://www.nist.gov/srd/nist1a.cfm
- SpectraBaseCorticosteronehttps://spectrabase.com/spectrum/GlEetH3l9I311.BETA.,21-DIHYDROXY-PREGN-4-ENE-3,20-DIONEhttps://spectrabase.com/spectrum/15fra6mtibFCorticosteronehttps://spectrabase.com/spectrum/1MTN7bBeJP3Corticosteronehttps://spectrabase.com/spectrum/6vSCWLl26LaCorticosteronehttps://spectrabase.com/spectrum/L335ZXOUJDMCORTICOSTERONhttps://spectrabase.com/spectrum/2EDKNSG9n0hCorticosteronehttps://spectrabase.com/spectrum/9jo4S1yEgCLCorticosteronehttps://spectrabase.com/spectrum/KpOWXT7EPJKCorticosteronehttps://spectrabase.com/spectrum/KKGUDA2syspCorticosteronehttps://spectrabase.com/spectrum/MKLYvjZCYxCorticosteronehttps://spectrabase.com/spectrum/DmKoHkybRTkCorticosteronehttps://spectrabase.com/spectrum/3ahdxJrcP8S
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlCompounds with biological roleshttp://www.genome.jp/kegg-bin/get_htext?br08001.keg
- LIPID MAPSLipid Classificationhttps://www.lipidmaps.org/
- Natural Product Activity and Species Source (NPASS)
- MassBank Europe
- Metabolomics Workbench
- Nature Synthesis
- PharosLICENSEData accessed from Pharos and TCRD is publicly available from the primary sources listed above. Please respect their individual licenses regarding proper use and redistribution.https://pharos.nih.gov/aboutcorticosteronehttps://pharos.nih.gov/ligands/CSXUDGJCZ8WB
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- Springer Nature
- SpringerMaterials
- The Cambridge Structural Database
- Wikidatacorticosteronehttps://www.wikidata.org/wiki/Q422543
- WikipediaCorticosteronehttps://en.wikipedia.org/wiki/Corticosterone
- Wiley
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmlCorticosteronehttps://www.ncbi.nlm.nih.gov/mesh/68003345Anti-Inflammatory Agentshttps://www.ncbi.nlm.nih.gov/mesh/68000893
- PubChem
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- EPA Substance Registry ServicesEPA SRS List Classificationhttps://sor.epa.gov/sor_internet/registry/substreg/LandingPage.do
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403415719https://pubchem.ncbi.nlm.nih.gov/substance/403415719
- NCBI