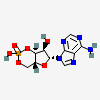
Adenosine cyclic phosphate
- Cyclic AMP
- cAMP
- 60-92-4
- Adenosine 3',5'-cyclic monophosphate
- 3',5'-cyclic AMP
- Create:2005-06-08
- Modify:2025-01-18
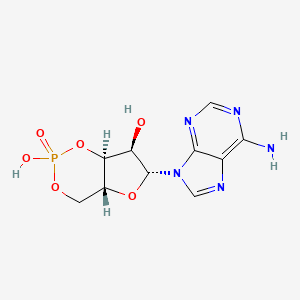
- 3',5'-Monophosphate, Adenosine Cyclic
- Adenosine Cyclic 3',5' Monophosphate
- Adenosine Cyclic 3',5'-Monophosphate
- Adenosine Cyclic 3,5 Monophosphate
- Adenosine Cyclic Monophosphate
- Adenosine Cyclic-3',5'-Monophosphate
- AMP, Cyclic
- Cyclic 3',5'-Monophosphate, Adenosine
- Cyclic AMP
- Cyclic AMP, (R)-Isomer
- Cyclic AMP, Disodium Salt
- Cyclic AMP, Monoammonium Salt
- Cyclic AMP, Monopotassium Salt
- Cyclic AMP, Monosodium Salt
- Cyclic AMP, Sodium Salt
- Cyclic Monophosphate, Adenosine
- Cyclic-3',5'-Monophosphate, Adenosine
- Monophosphate, Adenosine Cyclic
- Cyclic AMP
- cAMP
- 60-92-4
- Adenosine 3',5'-cyclic monophosphate
- 3',5'-cyclic AMP
- Adenosine 3',5'-cyclophosphate
- Adenosine 3',5'-phosphate
- cyclic 3',5'-AMP
- Adenosine cyclic monophosphate
- Cyclic Adenosine Monophosphate
- cyclic 3',5'-Adenylic acid
- Adenosine cyclophosphate
- Adenosine 3',5'-monophosphate
- Adenosine-3',5'-cyclophosphate
- cyclic Adenosine 3',5'-phosphate
- Adenosine cyclic 3',5'-monophosphate
- Adenosine, cyclic 3',5'-(hydrogen phosphate)
- Cyclic adenylic acid
- 3',5'-AMP
- Adenosine 3',5'-cyclic phosphate
- Adenosine cyclic 3',5'-phosphate
- cyclic Adenosine 3',5'-monophosphate
- CCRIS 4291
- Adenosine cyclic phosphate
- NSC 94017
- adenosine-3',5'-cyclic-monophosphate
- CHEBI:17489
- BRN 0052645
- [3H]cAMP
- [3H]-cAMP
- EINECS 200-492-9
- UNII-E0399OZS9N
- Adenosine 3',5'-phosphate monohydrate
- E0399OZS9N
- 3',5'-cAMP
- DTXSID8040436
- cyclic-AMP
- adenosine-3',5'-cyclic monophosphate
- adenosine-cyclic-phosphate
- adenosine cyclic-monophosphate
- 3' 5'-adenosine monophosphate
- CHEMBL316966
- adenosine-cyclic-phosphoric-acid
- (4aR,6R,7R,7aS)-6-(6-aminopurin-9-yl)-2-hydroxy-2-oxo-4a,6,7,7a-tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinin-7-ol
- 114697-05-1
- DTXCID6020436
- ADENOSINUM CYCLOPHOSPHORICUM
- 4-26-00-03618 (Beilstein Handbook Reference)
- MFCD00005845
- NSC-143670
- (4aR,6R,7R,7aS)-6-(6-amino-9H-purin-9-yl)-2,7-dihydroxy-hexahydro-1,3,5,2$l^{5}-furo[3,2-d][1,3,2$l^{5}]dioxaphosphinin-2-one
- adenosine 3',5'-(hydrogen phosphate)
- NCGC00093588-03
- Adenosine 3,5'-cyclic monophosphoric acid
- Adenosine cyclic 3',5'-(hydrogen phosphate)
- adenosine cyclic-3',5'-monophosphate
- 3',5'-Monophosphate, Adenosine Cyclic
- ADENOSINE CYCLIC-3',5'-(HYDROPHOSPHATE)
- ADENOSINE-3':5'-CYCLIC MONOPHOSPHORIC ACID (3:5-CYCLIC AMP)
- NSC-94017
- adenosine-3',5'-monophosphate
- AMP, Cyclic
- cyclic-3',5'-adenosine monophosphate
- Adenosine cyclicphosphate
- Adenosine-3',5'-cyclic phosphate
- Cyclic Monophosphate, Adenosine
- Monophosphate, Adenosine Cyclic
- CAS-60-92-4
- Adenosine Cyclic 3,5 Monophosphate
- Adenosine Cyclic 3',5' Monophosphate
- Cyclic 3',5'-Monophosphate, Adenosine
- Cyclic-3',5'-Monophosphate, Adenosine
- 3'5'-cyclic AMP
- Cyclic AMP (Standard)
- CYCLIC AMP; CAMP
- CYCLIC AMP [MI]
- Cyclic 3',5'-adenylate
- bmse000071
- Epitope ID:137352
- SCHEMBL1244
- Adenosini Cycloghosghas,(S)
- Lopac0_000084
- 3'5'-cyclic ester of AMP
- CYCLIC AMP [WHO-DD]
- GTPL2352
- GTPL5096
- cAMP; 3',5'-Cyclic AMP
- BDBM10851
- HY-B1511R
- cid_23902375
- HMS3260A10
- HY-B1511
- Tox21_113510
- Tox21_500084
- Adenosine-3',5'-cyclicmonophosphate
- BDBM50384264
- s9370
- Adenosine 3',5'-cyclophosphoric acid
- Adenosine 3':5'-cyclic monophosphate
- Adenosine 3,5'-cyclic monophosphorate
- Adenosine Cyclic 3':5'-Monophosphate
- AKOS015999561
- Tox21_113510_1
- 3', 5'-cyclic adenosine monophosphate
- Adenosine 3'-,5'-cyclic monophosphate
- CCG-204179
- DB02527
- LP00084
- SDCCGSBI-0050072.P002
- Adenosine 3a(2),5a(2)-cyclophosphate
- Adenosine-3',5'-cyclophosphate (cAMP)
- NCGC00093588-01
- NCGC00093588-02
- NCGC00093588-04
- NCGC00093588-06
- NCGC00260769-01
- AS-13112
- ADENOSINUM CYCLOPHOSPHORICUM [HPUS]
- Adenosine, 3',5'-cyclic-monophosphoric acid
- CS-0013299
- EU-0100084
- NS00122387
- ADENOSINE-3 ,5 -CYCLIC-MONOPHOSPHATE
- A 9501
- A51099
- C00575
- Q210041
- SR-01000075695
- CYCLIC-3',5'-(HYDROGEN PHOSPHATE)ADENOSINE
- SR-01000075695-1
- 4H-Furo[3,2-d]-1,3,2-dioxaphosphorin, adenosine deriv.
- B5C9F048-77DD-4170-A624-23C09E7B080D
- Adenosine 3',5'-cyclic monophosphate, >=98.5% (HPLC), powder
- Adenosine 3 inverted exclamation marka,5 inverted exclamation marka-cyclic monophosphate
- CAMP;Adenosine 3 inverted exclamation marka,5 inverted exclamation marka-cyclic monophosphat
- (1S,6R,8R,9R)-8-(6-aminopurin-9-yl)-3-hydroxy-3-oxo-2,4,7-trioxa-3$l^{5}-phosphabicyclo[4.3.0]nonan-9-ol
- (2R,4aR,6R,7R,7aS)-6-(6-amino-9H-purin-9-yl)tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide
- (4aR,6R,7R,7aS)-6-(6-amino-9H-purin-9-yl)-2,7-dihydroxy-hexahydro-1,3,5,2$l^{5}-furo[3,2-d][1,3,2]dioxaphosphinin-2-one
- (4aR,6R,7R,7aS)-6-(6-Amino-9H-purin-9-yl)-2,7-dihydroxytetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine 2-oxide
- (4aR,6R,7R,7aS)-6-(6-amino-9H-purin-9-yl)tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide
- (4aR,6R,7R,7aS)-6-(6-Aminopurin-9-yl)-2-oxo-tetrahydro-2-lambda*5*-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol
- 4H-furo[3,2-d]-1,3,2-dioxaphosphorin-7-ol, 6-(6-amino-9H-purin-9-yl)tetrahydro-2-hydroxy-, 2-oxide, (4aR,6R,7R,7aS)-
- 6-(6-Amino-9H-purin-9-yl)tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide #
176.8 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
172.6 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
188.6 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
181 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
188.8 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
171.8 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
172 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
172.6 Ų [M+H]+
172.7 Ų [M+H]+
189 Ų [M+Na]+
177 Ų [M+H]+
171.9 Ų [M-H]-
181.3 Ų [M+Na]+
182.3 Ų [M+Na]+
73.0 1
236.0 0.33
192.0 0.20
208.0 0.17
211.0 0.16
73.0 100
236.0 33.13
192.0 19.52
208.0 16.72
211.0 16.02
330.0603 100
136.0648 33.32
136.06352 100
330.05569 63.45
312.05301 8.82
119.03478 3.62
176.99254 2.71
330.0599 999
284.099482 163
331.062281 116
659.11108 71
171.149125 39
152.028412 999
134.07843 3
135.162689 2
212.187042 1
211.30896 1
134 100
133 17.68
79 11.19
78 3.24
134 100
133 17.24
79 12.63
- All Tissues
- Placenta
- Cytoplasm
- Extracellular
- Golgi apparatus
- Adenine phosphoribosyltransferase deficiency (APRT)
- Adenosine Deaminase Deficiency
- Adenylosuccinate Lyase Deficiency
- AICA-Ribosiduria
- Azathioprine Action Pathway
- Corticotropin Activation of Cortisol Production
- Dopamine Activation of Neurological Reward System
- Excitatory Neural Signalling Through 5-HTR 4 and Serotonin
- Excitatory Neural Signalling Through 5-HTR 6 and Serotonin
- Excitatory Neural Signalling Through 5-HTR 7 and Serotonin
- Total 36 pathways, visit the HMDB page for details
Information on 1 consumer products that contain cyclic AMP (adenosine cyclic phosphate) in the following categories is provided:
• Personal Care


H314 (16.7%): Causes severe skin burns and eye damage [Danger Skin corrosion/irritation]
H315 (50%): Causes skin irritation [Warning Skin corrosion/irritation]
H318 (16.7%): Causes serious eye damage [Danger Serious eye damage/eye irritation]
H319 (50%): Causes serious eye irritation [Warning Serious eye damage/eye irritation]
H335 (50%): May cause respiratory irritation [Warning Specific target organ toxicity, single exposure; Respiratory tract irritation]
P260, P261, P264, P264+P265, P271, P280, P301+P330+P331, P302+P352, P302+P361+P354, P304+P340, P305+P351+P338, P305+P354+P338, P316, P317, P319, P321, P332+P317, P337+P317, P362+P364, P363, P403+P233, P405, and P501
(The corresponding statement to each P-code can be found at the GHS Classification page.)
Aggregated GHS information provided per 6 reports by companies from 3 notifications to the ECHA C&L Inventory. Each notification may be associated with multiple companies.
Reported as not meeting GHS hazard criteria per 2 of 6 reports by companies. For more detailed information, please visit ECHA C&L website.
There are 2 notifications provided by 4 of 6 reports by companies with hazard statement code(s).
Information may vary between notifications depending on impurities, additives, and other factors. The percentage value in parenthesis indicates the notified classification ratio from companies that provide hazard codes. Only hazard codes with percentage values above 10% are shown.
Skin Corr. 1B (16.7%)
Skin Irrit. 2 (50%)
Eye Dam. 1 (16.7%)
Eye Irrit. 2 (50%)
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=IVOMOUWHDPKRLL-KQYNXXCUSA-N
- Australian Industrial Chemicals Introduction Scheme (AICIS)Adenosine, cyclic 3',5'-(hydrogen phosphate)https://services.industrialchemicals.gov.au/search-assessments/Adenosine, cyclic 3',5'-(hydrogen phosphate)https://services.industrialchemicals.gov.au/search-inventory/
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/
- ChemIDplusChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_useCyclic adenosine monophosphatehttps://www.drugbank.ca/drugs/DB02527
- EPA Chemicals under the TSCAAdenosine, cyclic 3',5'-(hydrogen phosphate)https://www.epa.gov/chemicals-under-tscaEPA TSCA Classificationhttps://www.epa.gov/tsca-inventory
- EPA DSSToxCompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-noticeAdenosine 3',5'-phosphate monohydratehttps://echa.europa.eu/substance-information/-/substanceinfo/100.000.448Adenosine 3',5'-phosphate monohydrate (EC: 200-492-9)https://echa.europa.eu/information-on-chemicals/cl-inventory-database/-/discli/details/63286
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linkingADENOSINE CYCLIC PHOSPHATEhttps://gsrs.ncats.nih.gov/ginas/app/beta/substances/E0399OZS9N
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingHMDB0000058_cms_306https://hmdb.ca/metabolites/HMDB0000058#spectra
- New Zealand Environmental Protection Authority (EPA)LICENSEThis work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International licence.https://www.epa.govt.nz/about-this-site/general-copyright-statement/Adenosine, cyclic 3',5'-(hydrogen phosphate)https://www.epa.govt.nz/industry-areas/hazardous-substances/guidance-for-importers-and-manufacturers/hazardous-substances-databases/
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/Cyclic ampNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBI3',5'-cyclic AMPhttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:17489
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/Cyclic AMPhttps://www.wikidata.org/wiki/Q210041LOTUS Treehttps://lotus.naturalproducts.net/
- NCI Thesaurus (NCIt)LICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuseNCI Thesaurushttps://ncit.nci.nih.gov
- Yeast Metabolome Database (YMDB)3',5'-cyclic AMPhttps://www.ymdb.ca/compounds/YMDB00023
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- Comparative Toxicogenomics Database (CTD)LICENSEIt is to be used only for research and educational purposes. Any reproduction or use for commercial purpose is prohibited without the prior express written permission of NC State University.http://ctdbase.org/about/legal.jsp
- Drug Gene Interaction database (DGIdb)LICENSEThe data used in DGIdb is all open access and where possible made available as raw data dumps in the downloads section.http://www.dgidb.org/downloadsCYCLIC ADENOSINE MONOPHOSPHATEhttps://www.dgidb.org/drugs/drugbank:DB02527COMPOUND 15A [PMID: 36642961]https://www.dgidb.org/drugs/iuphar.ligand:12352
- IUPHAR/BPS Guide to PHARMACOLOGYLICENSEThe Guide to PHARMACOLOGY database is licensed under the Open Data Commons Open Database License (ODbL) https://opendatacommons.org/licenses/odbl/. Its contents are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (http://creativecommons.org/licenses/by-sa/4.0/)https://www.guidetopharmacology.org/about.jsp#license[<sup>3</sup>H]cAMPhttps://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5096Guide to Pharmacology Target Classificationhttps://www.guidetopharmacology.org/targets.jsp
- Therapeutic Target Database (TTD)
- Consumer Product Information Database (CPID)LICENSECopyright (c) 2024 DeLima Associates. All rights reserved. Unless otherwise indicated, all materials from CPID are copyrighted by DeLima Associates. No part of these materials, either text or image may be used for any purpose other than for personal use. Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited without prior written permission.https://www.whatsinproducts.com/contents/view/1/6cyclic AMP (adenosine cyclic phosphate)https://www.whatsinproducts.com/chemicals/view/1/3018/000060-92-4Consumer Products Category Classificationhttps://www.whatsinproducts.com/
- ECI Group, LCSB, University of Luxembourg3',5'-cyclic AMP
- Natural Product Activity and Species Source (NPASS)Cyclic Adenosine Monophosphatehttps://bidd.group/NPASS/compound.php?compoundID=NPC212551
- EPA Chemical and Products Database (CPDat)EPA CPDat Classificationhttps://www.epa.gov/chemical-research/chemical-and-products-database-cpdat
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutcyclic-AMPhttps://foodb.ca/compounds/FDB030763
- MassBank EuropeAdenosine 3':5'-cyclicmonophosphatehttps://massbank.eu/MassBank/Result.jsp?inchikey=IVOMOUWHDPKRLL-KQYNXXCUSA-N
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-lawAdenosine 3',5'-cyclic monophosphatehttp://www.nist.gov/srd/nist1a.cfm
- SpectraBaseAdenosine 3',5'-cyclic monophosphatehttps://spectrabase.com/spectrum/AV7GXYbqYFLAdenosine 3',5'-cyclic monophosphatehttps://spectrabase.com/spectrum/8piiF8by8VbAdenosine-3',5'-cyclic phosphatehttps://spectrabase.com/spectrum/9rUAsMXqP5eAdenosine 3',5'-cyclic monophosphatehttps://spectrabase.com/spectrum/HTFkE5DNwfiADENOSINE-3',5'-CYCLOPHOSPHATEhttps://spectrabase.com/spectrum/C05XsNZFoaADENOSINE-3',5'-CYCLOPHOSPHATEhttps://spectrabase.com/spectrum/AyEYx4h2vU4
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlCompounds with biological roleshttp://www.genome.jp/kegg-bin/get_htext?br08001.keg
- MarkerDBLICENSEThis work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.https://markerdb.ca/Cyclic AMPhttps://markerdb.ca/chemicals/40
- Metabolomics Workbench
- National Drug Code (NDC) DirectoryLICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linkingADENOSINE CYCLIC PHOSPHATEhttps://www.fda.gov/drugs/drug-approvals-and-databases/national-drug-code-directory
- Nature Chemical Biology
- PharmGKBLICENSEPharmGKB data are subject to the Creative Commons Attribution-ShareALike 4.0 license (https://creativecommons.org/licenses/by-sa/4.0/).https://www.pharmgkb.org/page/policiescyclic adenosine monophosphatehttps://www.pharmgkb.org/chemical/PA166178640
- PharosLICENSEData accessed from Pharos and TCRD is publicly available from the primary sources listed above. Please respect their individual licenses regarding proper use and redistribution.https://pharos.nih.gov/about
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Springer Nature
- Wikidataadenosine cyclic phosphatehttps://www.wikidata.org/wiki/Q210041
- WikipediaCyclic adenosine monophosphatehttps://en.wikipedia.org/wiki/Cyclic_adenosine_monophosphate
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.html
- PubChem
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 389230984https://pubchem.ncbi.nlm.nih.gov/substance/389230984