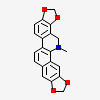
Dihydrosanguinarine
- Dihydrosanguinarine
- 3606-45-9
- Dihydro Sanguinarine
- 13,14-Dihydrosanguinarine
- dihydroavicine
- Create:2004-09-16
- Modify:2025-01-11
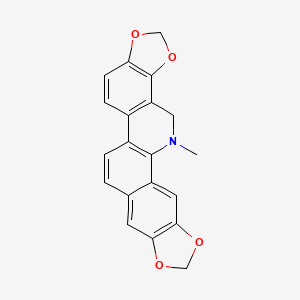
- dihydro-sanguinarine
- dihydrosanguinarine
- Dihydrosanguinarine
- 3606-45-9
- Dihydro Sanguinarine
- 13,14-Dihydrosanguinarine
- dihydroavicine
- dihydro-sanguinarine
- CHEBI:17209
- UNII-3H1ZKG80F7
- 3H1ZKG80F7
- 24-methyl-5,7,18,20-tetraoxa-24-azahexacyclo[11.11.0.02,10.04,8.014,22.017,21]tetracosa-1(13),2,4(8),9,11,14(22),15,17(21)-octaene
- DTXSID00189627
- 13,14-Dihydro-13-methyl-[1,3]benzodioxolo[5,6-c]-1,3-dioxolo[4,5-i]phenanthridine
- (1,3)Benzodioxolo(5,6-c)-1,3-dioxolo(4,5-i)phenanthridine, 13,14-dihydro-13-methyl-
- 13-methyl-13,14-dihydro-2H,10H-[1,3]dioxolo[4,5-i][1,3]dioxolo[4',5':4,5]benzo[1,2-c]phenanthridine
- Hydrosanguinarine
- 13, 14-dihydro-13-methyl-[1, 3]benzodioxolo[5, 6-c]-1, 3-dioxolo[4, 5-i]phenanthridine
- 3arv
- 3lle
- 13, 14-dihydro-13-methyl-(1, 3)benzodioxolo(5, 6-c)-1, 3-dioxolo(4, 5-i)phenanthridine
- 13,14-Dihydro-13-methyl-(1,3)benzodioxolo(5,6-c)-1,3-dioxolo(4,5-i)phenanthridine
- 13-methyl-13,14-dihydro-2H,10H-(1,3)dioxolo(4,5-i)(1,3)dioxolo(4',5':4,5)benzo(1,2-c)phenanthridine
- 24-methyl-5,7,18,20-tetraoxa-24-azahexacyclo(11.11.0.02,10.04,8.014,22.017,21)tetracosa-1(13),2,4(8),9,11,14(22),15,17(21)-octaene
- 3as0
- SCHEMBL420383
- CHEMBL465678
- DTXCID60112118
- HY-N0902
- BDBM50286637
- AKOS030526138
- CS-3819
- AC-34693
- DA-52553
- MS-25022
- 1ST179702
- NS00094596
- C05191
- F17675
- Q347588
- Dihydrosanguinarinedihydroavicine; Dihydro Sanguinarine
- 24-methyl-5,7,18,20-tetraoxa-24-azahexacyclo[11.11.0.0^{2,10.0^{4,8.0^{14,22.0^{17,21]tetracosa-1(13),2,4(8),9,11,14(22),15,17(21)-octaene
318.08062489167804 100
319.0875671794426 30.35
334.1081331787845 9.89
290.08278877409697 6.66
276.10557193163845 4.34
318.07647705078125 7352267
319.08428955078125 6071445
334.1075439453125 4582826.50
290.08154296875 1089100.12
304.09954833984375 1050243.50
Not Classified
Reported as not meeting GHS hazard criteria by 1 of 1 companies. For more detailed information, please visit ECHA C&L website.
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=CIUHLXZTZWTVFL-UHFFFAOYSA-N
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/Dihydrosanguinarinehttps://commonchemistry.cas.org/detail?cas_rn=3606-45-9
- ChemIDplusDihydrosanguinarinehttps://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0003606459ChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- EPA DSSToxDihydrosanguinarinehttps://comptox.epa.gov/dashboard/DTXSID00189627CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-noticeDihydrosanguinarinehttps://echa.europa.eu/substance-information/-/substanceinfo/100.235.388Dihydrosanguinarine (EC: 807-649-6)https://echa.europa.eu/information-on-chemicals/cl-inventory-database/-/discli/details/244528
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linkingDIHYDROSANGUINARINEhttps://gsrs.ncats.nih.gov/ginas/app/beta/substances/3H1ZKG80F7
- ChEBIDihydrosanguinarinehttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:17209
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/Dihydrosanguinarinehttps://www.wikidata.org/wiki/Q347588LOTUS Treehttps://lotus.naturalproducts.net/
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- Comparative Toxicogenomics Database (CTD)LICENSEIt is to be used only for research and educational purposes. Any reproduction or use for commercial purpose is prohibited without the prior express written permission of NC State University.http://ctdbase.org/about/legal.jspdihydrosanguinarinehttps://ctdbase.org/detail.go?type=chem&acc=C501843
- Crystallography Open Database (COD)LICENSEAll data in the COD and the database itself are dedicated to the public domain and licensed under the CC0 License. Users of the data should acknowledge the original authors of the structural data.https://creativecommons.org/publicdomain/zero/1.0/
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlPhytochemical compoundshttp://www.genome.jp/kegg-bin/get_htext?br08003.keg
- KNApSAcK Species-Metabolite Database
- Natural Product Activity and Species Source (NPASS)
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/DihydrosanguinarineNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics WorkbenchDihydrosanguinarinehttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=50814
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- SpectraBaseDIHYDROSANGUINARINEhttps://spectrabase.com/spectrum/BR09XIDtHb2DIHYDROSANGUINARINEhttps://spectrabase.com/spectrum/LhH2MRUtTKUDIHYDROSANGUINARINEhttps://spectrabase.com/spectrum/5EwkdkxXJwhSanguinarine artifact (dihydro-)https://spectrabase.com/spectrum/DkK3dhmLFAwDihydrosanguinarinehttps://spectrabase.com/spectrum/5wnAaFlEVdu
- Wikidatadihydrosanguinarinehttps://www.wikidata.org/wiki/Q347588
- WikipediaDigitoxigeninhttps://en.wikipedia.org/wiki/DigitoxigeninDihydrosanguinarinehttps://en.wikipedia.org/wiki/Dihydrosanguinarine
- PubChem
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmldihydrosanguinarinehttps://www.ncbi.nlm.nih.gov/mesh/67501843
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403167830https://pubchem.ncbi.nlm.nih.gov/substance/403167830
- NCBI