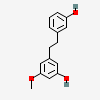
batatasin III
- batatasin III
- 56684-87-8
- 3-[2-(3-hydroxyphenyl)ethyl]-5-methoxyphenol
- 1-(3-Hydroxy-5-methoxyphenyl)-2-(3-hydroxyphenyl)ethane
- 3,3'-dihydroxy-5-methoxybibenzyl
- Create:2006-10-25
- Modify:2025-01-11
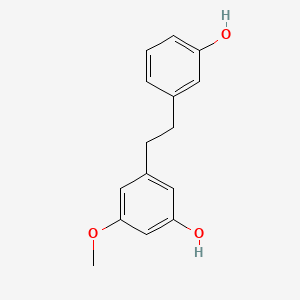
- batatasin III
- 56684-87-8
- 3-[2-(3-hydroxyphenyl)ethyl]-5-methoxyphenol
- 1-(3-Hydroxy-5-methoxyphenyl)-2-(3-hydroxyphenyl)ethane
- 3,3'-dihydroxy-5-methoxybibenzyl
- BatatasinIII
- Batatasin III (4)
- Phenol, 3-[2-(3-hydroxyphenyl)ethyl]-5-methoxy-
- CHEMBL450788
- SCHEMBL11505373
- DTXSID00904195
- CHEBI:174263
- BDBM246487
- EX-A6688
- AKOS028112322
- MS-23458
- DB-290684
- HY-122965
- CS-0090764
- NS00097305
- D85191
- 3-[2-(3-Hydroxyphenyl)ethyl]-5-methoxyphenol, 9CI
105.043221 100
121.063354 78.60
151.075043 73.12
103.054787 63.76
137.059021 61.89
106.0399 100
181.062 42.42
104.6539 39.19
185.0055 35.97
183.0691 34.45
106.0399 100
181.0620 42.42
104.6539 39.19
185.0055 35.97
183.0691 34.45
130.9664 100
227.0686 98.73
158.0688 40.17
136.0506 34.82
183.0844 23.39
243.1022491455078 3056408.25
227.07077026367188 632918.19
228.0789794921875 98602.30
122.03600311279297 63758.04
201.09129333496094 33125.94
245.1168670654297 100
121.06454467773438 88.58
151.07518005371094 55.19
137.05953979492188 42.15
93.06966400146484 6.98
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=VYQXIUVIYICVCM-UHFFFAOYSA-N
- BindingDBLICENSEAll data curated by BindingDB staff are provided under the Creative Commons Attribution 3.0 License (https://creativecommons.org/licenses/by/3.0/us/).https://www.bindingdb.org/rwd/bind/info.jsp
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/
- EPA DSSToxBatatasin IIIhttps://comptox.epa.gov/dashboard/DTXSID00904195CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingBatatasin IIIhttp://www.hmdb.ca/metabolites/HMDB0030636HMDB0030636_msms_452122https://hmdb.ca/metabolites/HMDB0030636#spectra
- ChEBI
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/batatasin IIIhttps://www.wikidata.org/wiki/Q82873435LOTUS Treehttps://lotus.naturalproducts.net/
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- Japan Chemical Substance Dictionary (Nikkaji)
- KNApSAcK Species-Metabolite Database
- Natural Product Activity and Species Source (NPASS)
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics Workbench
- SpectraBaseBATATASIN-III;3,3'-DIHYDROXY-ETHANEDIYL-5-METHOXYDIBENZYLhttps://spectrabase.com/spectrum/HI0MsFgI4JaBATATASIN 3https://spectrabase.com/spectrum/6JQvPgVSyDk
- Springer Nature
- Wikidatabatatasin IIIhttps://www.wikidata.org/wiki/Q82873435
- PubChem
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/NORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403029437https://pubchem.ncbi.nlm.nih.gov/substance/403029437