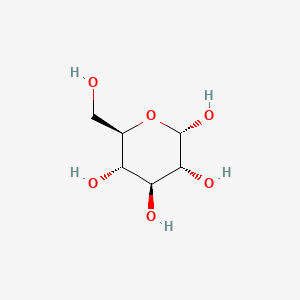
alpha-D-Glucopyranose
- alpha-D-glucose
- alpha-D-Glucopyranose
- 492-62-6
- alpha-Dextrose
- alpha-glucose
- Create:2004-09-16
- Modify:2025-01-18

- 1,3-alpha-D-glucan
- alpha-1,3-glucan
- alpha-D-glucose
- alpha-D-Glucopyranose
- 492-62-6
- alpha-Dextrose
- alpha-glucose
- (2S,3R,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol
- Glucopyranose, alpha-D-
- alpha-D-Glc
- (2S,3R,4S,5S,6R)-6-(Hydroxymethyl)tetrahydro-2H-pyran-2,3,4,5-tetraol
- CHEBI:17925
- glucoses
- Grape sugar
- Corn sugar
- 5J5I9EB41E
- CHEMBL423707
- D-gluose
- a-D-Glucopyranose
- MFCD00063774
- .alpha.-D-Glucopyranose
- alpha-D-glucose, Phase I
- alpha-D-glucose, Phase II
- (1,4-alpha-D-Glucosyl)n
- alpha-D-glucopyranose, Phase I
- D-gluco-hexose
- alpha-D-glucopyranose, Phase II
- a-D-Glucose
- .alpha.-D-Glucose
- (+)-Glucose
- D-Glucose-5-C-tD-glucose [MeSH: Glucose]
- GLUCOSE, ALPHA-D-
- UNII-5J5I9EB41E
- a-Dextrose
- GLUCOSE, ANHYDROUS [WHO-IP]
- a-Glucose
- alpha-delta-Glucose
- EINECS 207-757-8
- alpha-d(+)-glucose
- I+/--D-Glucose
- 1,3-alpha-D-Glucan
- 1,4-alpha-D-Glucan
- 1,6-alpha-D-Glucan
- |A-D-Glucose anhydrous
- alpha-delta-Glucopyranose
- D(+)Glucose, Anhydrous
- Dextrin from corn, p.a.
- (2S,3R,4S,5S,6R)-6-(hydroxymethyl)tetrahydropyran-2,3,4,5-tetrol
- bmse000015
- bmse000791
- bmse000797
- bmse000855
- Epitope ID:144998
- MolMap_000023
- SCHEMBL6222
- (1->3)-alpha-D-glucan
- (1->4)-alpha-D-glucan
- (1->6)-alpha-D-glucan
- (1,6-alpha-D-Glucosyl)n
- MLS006011570
- Glucose, p.a., ACS reagent
- (1->3)-alpha-D-glucopyranan
- (1->4)-alpha-D-glucopyranan
- (1,4-alpha-D-Glucosyl)n+1
- (1,4-alpha-D-Glucosyl)n-1
- [alpha-D-Glucosyl-(1,3)]n
- CHEBI:15444
- CHEBI:18269
- CHEBI:18398
- CHEBI:28100
- CHEBI:28102
- D-(+)-Glucose, AR, anhydrous
- DTXSID30197710
- [alpha-D-Glucosyl-(1,3)]n+1
- alpha-D-Glucose, anhydrous, 96%
- CMC_6867
- Dextrin from corn, Type I, powder
- D-(+) Glucose, analytical standard
- D-(+)-Glucose, analytical standard
- alpha-D-glucose; D-glucose; glucose
- BDBM50351158
- s6028
- AKOS015950677
- D-(+)-Glucose, >=99% (GC)
- NCGC00160621-01
- NCGC00160621-03
- NCGC00160621-05
- NCGC00160621-08
- 27707-45-5
- AC-15067
- BS-17112
- SMR004703328
- D-(+)-Glucose, >=99.5% (GC)
- GLUCOSUM, ANHYDROUS [WHO-IP LATIN]
- HY-128417
- CS-0099249
- NS00074067
- D-(+)-Glucose, LR, anhydrous, >=99.5%
- D-(+)-Glucose, tested according to Ph.Eur.
- EN300-59169
- alpha-D-Glucose, SAJ first grade, >=98.0%
- C00267
- D-(+)-Glucose, BioXtra, >=99.5% (GC)
- D70945
- 4-{(1,4)-alpha-D-Glucosyl}(n-1)-D-glucose
- alpha-D-Glucose, SAJ special grade, >=98.0%
- D-Glucose (Dextrose), NIST(R) SRM(R) 917C
- W-204032
- BRD-K87424134-001-01-0
- D-(+)-Glucose, plant cell culture tested, BioReagent
- D-(+)-Glucose, Vetec(TM) reagent grade, >=99.5%
- Dextrin from corn, commercial grade, Type II, powder
- Q23905965
- Dextrose, meets EP, BP, JP, USP testing specifications
- Glucose, European Pharmacopoeia (EP) Reference Standard
- Z905052654
- 074AD9E3-1FC7-485C-8A50-2B653D501E5B
- D-(+)-Glucose, suitable for mouse embryo, >=99.5% (GC)
- D-(+)-Glucose, 99.9 atom % 16O, 99.9 atom % 12C
- Dextrose, United States Pharmacopeia (USP) Reference Standard
- D-(+)-Glucose, anhydrous, free-flowing, Redi-Dri(TM), >=99.5%
- Dextrose, meets EP, BP, JP, USP testing specifications, anhydrous
- D-(+)-Glucose, BioUltra, anhydrous, >=99.5% (sum of enantiomers, HPLC)
- D-(+)-Glucose, Hybri-Max(TM), powder, BioReagent, suitable for hybridoma
- D-(+)-Glucose, meets analytical specification of Ph. Eur., BP, anhydrous
- Dextrose, Pharmaceutical Secondary Standard; Certified Reference Material
- D-(+)-Glucose, powder, BioReagent, suitable for cell culture, suitable for insect cell culture, suitable for plant cell culture, >=99.5%
147.1 Ų [M+Na]+
146.5 Ų [M+Na]+
85.0 100
73.0 25.98
93.0 16.16
117.0 15.87
75.0 10.36
85.0 100
73.0 78.77
41.0 39.27
45.0 29.75
61.0 21.29
59.01363 100
71.01395 78.20
89.02454 31.90
101.02419 19.40
85.02976 11.10
59.01381 100
59.02263 37.50
71.0137 16.20
71.02491 8.20
43.01873 5
204.099823 100
191.09198 90.40
281.193848 17.38
192.090744 11.31
140.031738 9.51
204.09935 100
191.091537 57.75
189.075974 13.19
147.06543 12.76
73.046799 11.91
- Epidermis
- Intestine
- Fanconi-bickel syndrome
- Fructose and mannose metabolism
- Fructose intolerance, hereditary
- Fructose-1,6-diphosphatase deficiency
- Fructosuria
- Galactose Metabolism
- Galactosemia
- Galactosemia II (GALK)
- Galactosemia III
- Glycogen Storage Disease Type 1A (GSD1A) or Von Gierke Disease
- Total 28 pathways, visit the HMDB page for details
Not Classified
Reported as not meeting GHS hazard criteria by 48 of 49 companies (only 2% companies provided GHS information). For more detailed information, please visit ECHA C&L website.
Aggregated GHS information provided per 49 reports by companies from 3 notifications to the ECHA C&L Inventory.
Reported as not meeting GHS hazard criteria per 48 of 49 reports by companies. For more detailed information, please visit ECHA C&L website.
There is 1 notification provided by 1 of 49 reports by companies with hazard statement code(s).
Information may vary between notifications depending on impurities, additives, and other factors. The percentage value in parenthesis indicates the notified classification ratio from companies that provide hazard codes. Only hazard codes with percentage values above 10% are shown.
IMAP assessments - .alpha.-D-Glucopyranose: Human health tier I assessment
IMAP assessments - .alpha.-D-Glucopyranose: Environment tier I assessment
PubMed: 28270806
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
PubMed: 6589104, 16277678, 15338487, 10361015, 15249323
Tie-juan ShaoZhi-xing HeZhi-jun XieHai-chang LiMei-jiao WangCheng-ping Wen. Characterization of ankylosing spondylitis and rheumatoid arthritis using 1H NMR-based metabolomics of human fecal extracts. Metabolomics. April 2016, 12:70: https://link.springer.com/article/10.1007/s11306-016-1000-2
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=WQZGKKKJIJFFOK-DVKNGEFBSA-N
- Australian Industrial Chemicals Introduction Scheme (AICIS).alpha.-D-Glucopyranosehttps://services.industrialchemicals.gov.au/search-assessments/.alpha.-D-Glucopyranosehttps://services.industrialchemicals.gov.au/search-inventory/
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/α-D-Glucopyranose, homopolymerhttps://commonchemistry.cas.org/detail?cas_rn=27707-45-5
- ChemIDplusChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- EPA DSSToxalpha-D-Glucopyranosehttps://comptox.epa.gov/dashboard/DTXSID30197710CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-noticeα-D-glucose (EC: 207-757-8)https://echa.europa.eu/information-on-chemicals/cl-inventory-database/-/discli/details/95132
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linking.ALPHA.-D-GLUCOPYRANOSEhttps://gsrs.ncats.nih.gov/ginas/app/beta/substances/5J5I9EB41E
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingalpha-D-Glucosehttp://www.hmdb.ca/metabolites/HMDB0003345HMDB0003345_msms_2254https://hmdb.ca/metabolites/HMDB0003345#spectra
- New Zealand Environmental Protection Authority (EPA)LICENSEThis work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International licence.https://www.epa.govt.nz/about-this-site/general-copyright-statement/
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/DextroseNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBI
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/(2S,3R,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrolhttps://www.wikidata.org/wiki/Q23905965LOTUS Treehttps://lotus.naturalproducts.net/
- Yeast Metabolome Database (YMDB)Alpha-D-Glucosehttps://www.ymdb.ca/compounds/YMDB00273
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- Crystallography Open Database (COD)LICENSEAll data in the COD and the database itself are dedicated to the public domain and licensed under the CC0 License. Users of the data should acknowledge the original authors of the structural data.https://creativecommons.org/publicdomain/zero/1.0/
- ECI Group, LCSB, University of Luxembourgalpha-D-glucose
- Natural Product Activity and Species Source (NPASS)Alpha-D-Glucosehttps://bidd.group/NPASS/compound.php?compoundID=NPC92246
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutPolydextrosehttps://foodb.ca/compounds/FDB010093alpha-D-Glucopyranosehttps://foodb.ca/compounds/FDB011829
- GlyCosmos Glycoscience PortalLICENSEAll copyrightable parts of the datasets in GlyCosmos are under the Creative Commons Attribution (CC BY 4.0) License.https://glycosmos.org/license
- GlyGen
- Haz-Map, Information on Hazardous Chemicals and Occupational DiseasesLICENSECopyright (c) 2022 Haz-Map(R). All rights reserved. Unless otherwise indicated, all materials from Haz-Map are copyrighted by Haz-Map(R). No part of these materials, either text or image may be used for any purpose other than for personal use. Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited without prior written permission.https://haz-map.com/Aboutalpha-D-Glucosehttps://haz-map.com/Agents/11730
- SpectraBasealpha-D-Glucosehttps://spectrabase.com/spectrum/GBRbq5nrbGOalpha GLUCOSEhttps://spectrabase.com/spectrum/4QFiFFWXlfsalpha(D) GLUCOPYRANOSEhttps://spectrabase.com/spectrum/7VjYuWGXAKWALPHA-D-GLUCOPYRANOSEhttps://spectrabase.com/spectrum/I6dYQB94drMα-D-Glucosehttps://spectrabase.com/spectrum/InxwwCCdyYUalpha-D-Glucosehttps://spectrabase.com/spectrum/LuiHqanULRPGlucose monohydratehttps://spectrabase.com/spectrum/Hdvol7UjQvgalpha-D-Glucosehttps://spectrabase.com/spectrum/Ko8Q5mMhpMPα-D-Glucosehttps://spectrabase.com/spectrum/8ettxwkd78falpha-D-Glucosehttps://spectrabase.com/spectrum/78OQcVbr5mG
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.html
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics Workbench
- Nature Chemical Biology
- NCI Thesaurus (NCIt)LICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuseNCI Thesaurushttps://ncit.nci.nih.gov
- NMRShiftDB
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- Springer Nature
- SpringerMaterials
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- Wikidataα-D-glucopyranosehttps://www.wikidata.org/wiki/Q23905965
- Wiley
- PubChem
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmlalpha-1,3-glucanhttps://www.ncbi.nlm.nih.gov/mesh/67045788
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- Glycan Naming and Subsumption Ontology (GNOme)GNOme
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403391667https://pubchem.ncbi.nlm.nih.gov/substance/403391667

 D-Glucose
D-Glucose Dextrin
Dextrin







