Pyridoxal phosphate
- pyridoxal phosphate
- 54-47-7
- Codecarboxylase
- pyridoxal 5-phosphate
- pyridoxal 5'-phosphate
- Create:2004-09-16
- Modify:2025-02-01
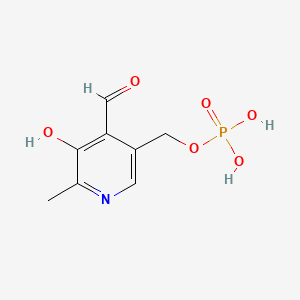
- Phosphate, Pyridoxal
- Pyridoxal 5 Phosphate
- Pyridoxal 5-Phosphate
- Pyridoxal P
- Pyridoxal Phosphate
- Pyridoxal-P
- pyridoxal phosphate
- 54-47-7
- Codecarboxylase
- pyridoxal 5-phosphate
- pyridoxal 5'-phosphate
- Pyridoxal P
- Pyridoxyl phosphate
- (4-formyl-5-hydroxy-6-methylpyridin-3-yl)methyl dihydrogen phosphate
- Biosechs
- Hairoxal
- Pidopidon
- Pyromijin
- Vitazechs
- Pyridoxal-5'-phosphate
- Hiadelon
- Himitan
- Phosphopyridoxal
- Sechvitan
- Pydoxal
- Piodel
- Apolon B6
- HI-Pyridoxin
- 853645-22-4
- Pal-P
- PYRIDOXAL-5-PHOSPHATE
- pyridoxal-P
- Phosphopyridoxal coenzyme
- Vitahexin P
- Hexermin P
- Coenzyme B6
- Pyridoxal monophosphate
- Pyridoxaldehyde phosphate
- Apolon B(sub 6)
- Phosphoridoxal coenzyme
- PLP
- pyridoxal-5P
- Pyridoxal 5'-(dihydrogen phosphate)
- Pyridoxal 5-monophosphoric acid ester
- Vitamin B6 phosphate
- Vitahexin-P
- Hexermin-P
- Pyridoxal phosphate [JAN]
- 3-Hydroxy-2-methyl-5-((phosphonooxy)methyl)-4-pyridinecarboxaldehyde
- Pyridoxal phosphate anhydrous
- 4-Pyridinecarboxaldehyde, 3-hydroxy-2-methyl-5-((phosphonooxy)methyl)-
- 3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]-4-pyridinecarboxaldehyde
- Pyridoxal 5/'-phosphate (hydrate)
- Pyridoxal, 5-(dihydrogen phosphate)
- MFCD00006333
- NSC-82388
- 3-Hydroxy-5-(hydroxymethyl)-2-methylisonicotinaldehyde 5-phosphate
- 2-Methyl-3-hydroxy-4-formyl-5-hydroxymethylpyridine-5-calcium phosphate
- 4-pyridinecarboxaldehyde, 3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]-
- CHEMBL82202
- F06SGE49M6
- CHEBI:18405
- pyridoxal-phosphate
- 3-Hydroxy-2-methyl-5-([phosphonooxy]methyl)-4-pyridinecarboxaldehyde
- NSC82388
- MC-1
- [(4-formyl-5-hydroxy-6-methylpyridin-3-yl)methoxy]phosphonic acid
- 4-Pyridinecarboxaldehyde,3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]-
- 2-Methyl-3-hydroxy-4-formyl-5-hydroxymethylpyridine-5-calcium phosphate trihydrate
- (4-formyl-5-hydroxy-6-methyl-3-pyridyl)methyl dihydrogen phosphate
- Pyridoxalphosphate
- Pyridoxal 5'-phosphate 5
- C8H10NO6P
- Pyridoxal 5'-phosphate;Pyridoxyl phosphate
- Pyridoxal-5-Phosphate Hydrate
- Sechvitan, Vitahexin P
- NCGC00166300-01
- Pyridoxal-5-monophosphate
- EINECS 200-208-3
- NSC 82388
- Pyridoxal phosphate (6CI)
- UNII-F06SGE49M6
- Vitamin B6 phosphate (ester)
- Pyridoxal, 5-(dihydrogenphosphate)
- SRI 2392
- P-5'-P
- Pridoxal-5-Phosphate
- Pyridoxal 5 inverted exclamation marka-phosphate hydrate
- pyridoxal 5''-phosphate
- P5P
- Pyridoxal, 5-(dihydrogen phosphate) (8CI)
- (4-Formyl-5-hydroxy-6-methyl(3-pyridyl))methyl dihydrogen phosphate
- bmse000111
- Pyridoxal 5'-phosphate monohydrate, vitamin B6
- SCHEMBL23158
- Pyridoxal phosphate (Standard)
- GTPL5249
- SGCUT00188
- DTXSID4048351
- HY-B1744R
- Pyridoxal 5'-phosphate anhydrous
- 2-Methyl-3-hydroxy-4-formyl-5-pyridylmethylphosphoric acid
- EX-A980
- 4-Formyl-5-hydroxy-6-methyl-pyridin-3-yl)methoxyphosphonic acid
- BCP34576
- HY-B1744
- PYRIDOXAL 5-PHOSPHATE [MI]
- Pyridoxal phosphate treated .beta.-lactoglobulin from bovine whey
- to_000077
- BDBM50118216
- PYRIDOXAL PHOSPHATE [WHO-DD]
- s5311
- STL185213
- pyridoxal 5''-(dihydrogen phosphate)
- AKOS015891654
- Phosphoric acid mono-(4-formyl-5-hydroxy-6-methyl-pyridin-3-ylmethyl) ester
- Pyridoxal 5'-phosphate;Codecarboxylase
- CCG-266929
- CS-7767
- DB00114
- SB18794
- PYRIDOXAL 5'-PHOSPHATE [VANDF]
- Pyridoxal 5'-phosphate hydrate, >=98%
- AS-19314
- Pyridoxal 5-phosphate;Pyridoxyl phosphate
- SY065874
- DB-052584
- VITAMIN B6 (PYRIDOXAL 5-PHOSPHATE)
- Isonicotinaldehyde, 5-(dihydrogen phosphate)
- NS00014024
- C00018
- F17391
- SBI-0633616.0002
- EN300-6474442
- Pyridoxal 5'-phosphate monohydrate - Vitamin B6
- Q418957
- SR-01000944534
- SR-01000944534-1
- BRD-K50712474-001-02-7
- BRD-K50712474-002-02-5
- A26BDB6A-282A-4D13-A916-7B2B215B0FD6
- Z1741970251
- (4-Formyl-5-hydroxy-6-methylpyridin-3-yl)methyldihydrogenphosphate
- 4-Pyridinecarboxaldehyde, 3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]- (9CI)
- Pyridoxal 5'-phosphate hydrate, powder, BioReagent, suitable for cell culture
160.8 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
149.4 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
151.3 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
153.2 Ų [M+Na-2H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
161.4 Ų [M+Na]+ [CCS Type: DT; Method: stepped-field]
150.8 Ų [M-H]- [CCS Type: DT; Method: stepped-field]
151.94 Ų [M+H]+ [CCS Type: DT; Method: stepped-field]
146.34 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with Agilent tune mix (Agilent)]
151.4 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with Agilent tune mix (Agilent)]
165.05 Ų [M+Na]+ [CCS Type: DT; Method: stepped-field]
154.76 Ų [M+H]+ [CCS Type: DT; Method: stepped-field]
150.54 Ų [M-H]- [CCS Type: DT; Method: stepped-field]
150.1 Ų [M-H]-
161.4 Ų [M+Na]+
162.4 Ų [M+Na]+
152.4 Ų [M+H]+
151.7 Ų [M+H]+
149.4 Ų [M-H]-
153.2 Ų [M-2H+Na]-
219.0 1
251.0 0.67
211.0 0.55
243.0 0.51
133.0 0.40
178.0 100
147.0 89.39
179.0 80.48
119.0 58.96
211.0 52.25
248.0 100
150.0 68.29
122.0 3.14
168.0 3.11
94.0 1.71
94.0 100
150.0 94.86
122.0 58.83
67.0 53.04
106.0 19.80
246.3 999
164.4 80
96.9 55
228.4 14
79 11
164.2 999
97 922
79 181
246.1 68
146.9 63
150 100
122 15.93
94 8.84
93 6.81
149 4.32
 Pyridoxal (annotation moved to)
Pyridoxal (annotation moved to)- Erythrocyte
- Kidney
- Liver
- Neuron
- Skeletal Muscle
- 2-aminoadipic 2-oxoadipic aciduria
- 2-Hydroxyglutric Aciduria (D And L Form)
- 2-Methyl-3-Hydroxybutryl CoA Dehydrogenase Deficiency
- 3-Hydroxy-3-Methylglutaryl-CoA Lyase Deficiency
- 3-hydroxyisobutyric acid dehydrogenase deficiency
- 3-hydroxyisobutyric aciduria
- 3-Methylcrotonyl Coa Carboxylase Deficiency Type I
- 3-Methylglutaconic Aciduria Type I
- 3-Methylglutaconic Aciduria Type III
- 3-Methylglutaconic Aciduria Type IV
- Total 137 pathways, visit the HMDB page for details
PubMed: 6589104, 16277678, 15338487, 10361015, 15249323
Tie-juan ShaoZhi-xing HeZhi-jun XieHai-chang LiMei-jiao WangCheng-ping Wen. Characterization of ankylosing spondylitis and rheumatoid arthritis using 1H NMR-based metabolomics of human fecal extracts. Metabolomics. April 2016, 12:70: https://link.springer.com/article/10.1007/s11306-016-1000-2
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=NGVDGCNFYWLIFO-UHFFFAOYSA-N
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/Pyridoxal 5′-phosphatehttps://commonchemistry.cas.org/detail?cas_rn=54-47-7
- ChemIDplusPyridoxal phosphatehttps://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0000054477ChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_usePyridoxal phosphatehttps://www.drugbank.ca/drugs/DB00114
- DTP/NCILICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuse
- EPA DSSToxPyridoxal phosphatehttps://comptox.epa.gov/dashboard/DTXSID4048351CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linkingPYRIDOXAL PHOSPHATE ANHYDROUShttps://gsrs.ncats.nih.gov/ginas/app/beta/substances/F06SGE49M6
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingPyridoxal 5'-phosphatehttp://www.hmdb.ca/metabolites/HMDB0001491HMDB0001491_cms_31340https://hmdb.ca/metabolites/HMDB0001491#spectra
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/MAGNESIUM PYRIDOXAL 5-PHOSPHATE GLUTAMATENORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBIPyridoxal 5'-phosphatehttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:18405
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/Pyridoxal phosphatehttps://www.wikidata.org/wiki/Q418957LOTUS Treehttps://lotus.naturalproducts.net/
- NCI Thesaurus (NCIt)LICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuse
- Open TargetsLICENSEDatasets generated by the Open Targets Platform are freely available for download.https://platform-docs.opentargets.org/licencePYRIDOXAL PHOSPHATE ANHYDROUShttps://platform.opentargets.org/drug/CHEMBL82202
- Yeast Metabolome Database (YMDB)Pyridoxal 5'-phosphatehttps://www.ymdb.ca/compounds/YMDB00363
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- ClinicalTrials.govLICENSEThe ClinicalTrials.gov data carry an international copyright outside the United States and its Territories or Possessions. Some ClinicalTrials.gov data may be subject to the copyright of third parties; you should consult these entities for any additional terms of use.https://clinicaltrials.gov/ct2/about-site/terms-conditions#Use
- Comparative Toxicogenomics Database (CTD)LICENSEIt is to be used only for research and educational purposes. Any reproduction or use for commercial purpose is prohibited without the prior express written permission of NC State University.http://ctdbase.org/about/legal.jspPyridoxal Phosphatehttps://ctdbase.org/detail.go?type=chem&acc=D011732
- Therapeutic Target Database (TTD)Pyridoxal Phosphatehttps://idrblab.net/ttd/data/drug/details/D06JGH
- DailyMedPYRIDOXAL PHOSPHATE ANHYDROUShttps://dailymed.nlm.nih.gov/dailymed/search.cfm?labeltype=all&query=PYRIDOXAL+PHOSPHATE+ANHYDROUS
- ECI Group, LCSB, University of Luxembourgpyridoxal 5'-phosphate
- KNApSAcK Species-Metabolite Database
- Natural Product Activity and Species Source (NPASS)Pyridoxal Phosphatehttps://bidd.group/NPASS/compound.php?compoundID=NPC36498
- EPA Chemical and Products Database (CPDat)Pyridoxal phosphatehttps://comptox.epa.gov/dashboard/DTXSID4048351#exposureEPA CPDat Classificationhttps://www.epa.gov/chemical-research/chemical-and-products-database-cpdat
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutPyridoxal 5'-phosphatehttps://foodb.ca/compounds/FDB021820
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-lawPyridoxal 5-phosphatehttp://www.nist.gov/srd/nist1a.cfm
- SpectraBasePyridoxal-5-phosphatehttps://spectrabase.com/spectrum/ErhsJuRu8FPyridoxal-5-phosphatehttps://spectrabase.com/spectrum/AFEa8wzC4I3Pyridoxal 5'-phosphatehttps://spectrabase.com/spectrum/FA3j9JG7ll8
- IUPAC Digitized pKa DatasetPyridoxal 5-phosphatehttps://github.com/IUPAC/Dissociation-Constants
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlCompounds with biological roleshttp://www.genome.jp/kegg-bin/get_htext?br08001.kegAnatomical Therapeutic Chemical (ATC) classificationhttp://www.genome.jp/kegg-bin/get_htext?br08303.keg
- MarkerDBLICENSEThis work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.https://markerdb.ca/Pyridoxal 5'-phosphatehttps://markerdb.ca/chemicals/631
- MassBank EuropePyridoxal 5-phosphatehttps://massbank.eu/MassBank/Result.jsp?inchikey=NGVDGCNFYWLIFO-UHFFFAOYSA-N
- Metabolomics WorkbenchPyridoxal 5'-phosphatehttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=37824
- National Drug Code (NDC) DirectoryLICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linkingPYRIDOXAL PHOSPHATE ANHYDROUShttps://www.fda.gov/drugs/drug-approvals-and-databases/national-drug-code-directory
- Nature Chemical Biology
- NIPH Clinical Trials Search of Japan
- PharmGKBLICENSEPharmGKB data are subject to the Creative Commons Attribution-ShareALike 4.0 license (https://creativecommons.org/licenses/by-sa/4.0/).https://www.pharmgkb.org/page/policiespyridoxal phosphatehttps://www.pharmgkb.org/chemical/PA164749650
- PharosLICENSEData accessed from Pharos and TCRD is publicly available from the primary sources listed above. Please respect their individual licenses regarding proper use and redistribution.https://pharos.nih.gov/aboutpyridoxal phosphatehttps://pharos.nih.gov/ligands/K6BKDU9BHL46
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Springer Nature
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- WHO Anatomical Therapeutic Chemical (ATC) ClassificationLICENSEUse of all or parts of the material requires reference to the WHO Collaborating Centre for Drug Statistics Methodology. Copying and distribution for commercial purposes is not allowed. Changing or manipulating the material is not allowed.https://www.whocc.no/copyright_disclaimer/Pyridoxal phosphatehttps://www.whocc.no/atc_ddd_index/?code=A11HA06
- Wikidatapyridoxal phosphatehttps://www.wikidata.org/wiki/Q418957
- WikipediaPyridoxal phosphatehttps://en.wikipedia.org/wiki/Pyridoxal_phosphate
- Wiley
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmlPyridoxal Phosphatehttps://www.ncbi.nlm.nih.gov/mesh/68011732Vitamin B Complexhttps://www.ncbi.nlm.nih.gov/mesh/68014803
- PubChem
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- EPA Substance Registry ServicesEPA SRS List Classificationhttps://sor.epa.gov/sor_internet/registry/substreg/LandingPage.do
- EPA Chemicals under the TSCAEPA TSCA Classificationhttps://www.epa.gov/tsca-inventory
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403383870https://pubchem.ncbi.nlm.nih.gov/substance/403383870
- NCBI














