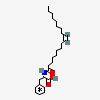
N-oleoyl phenylalanine
PubChem CID
52922059
Molecular Formula
Synonyms
- N-oleoyl phenylalanine
- N-oleoyl-L-phenylalanine
- 136560-78-6
- oleoyl-L-Phe-OH
- N-(9Z-octadecenoyl)-phenylalanine
Molecular Weight
429.6 g/mol
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Dates
- Create:2011-06-09
- Modify:2025-01-11
Description
N-oleoyl-L-phenylalanine is an N-acyl-L-phenylalanine resulting from the formal condensation of the carboxy group of oleic acid with the amino group of L-phenylalanine. It is a N-acyl-L-phenylalanine and a fatty amide. It is functionally related to an oleic acid. It is a conjugate acid of a N-oleoyl-L-phenylalaninate.
Chemical Structure Depiction
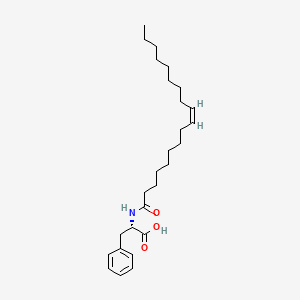
Conformer generation is disallowed since too flexible
SVG Image

IUPAC Condensed
oleoyl-Phe-OH
Sequence
F
HELM
PEPTIDE1{[CCCCCCCC/C=C\CCCCCCCC(=O)N[C@@H](Cc1ccccc1)C(=O)O]}$$$$
IUPAC
N-oleoyl-L-phenylalanine
(2S)-2-[[(Z)-octadec-9-enoyl]amino]-3-phenylpropanoic acid
Computed by Lexichem TK 2.7.0 (PubChem release 2021.05.07)
InChI=1S/C27H43NO3/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-19-22-26(29)28-25(27(30)31)23-24-20-17-16-18-21-24/h9-10,16-18,20-21,25H,2-8,11-15,19,22-23H2,1H3,(H,28,29)(H,30,31)/b10-9-/t25-/m0/s1
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
UWKNPULCJWBBDD-JRUKXMRZSA-N
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
CCCCCCCC/C=C\CCCCCCCC(=O)N[C@@H](CC1=CC=CC=C1)C(=O)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C27H43NO3
Computed by PubChem 2.1 (PubChem release 2021.05.07)
136560-78-6
- N-oleoyl phenylalanine
- N-oleoyl-L-phenylalanine
- 136560-78-6
- oleoyl-L-Phe-OH
- N-(9Z-octadecenoyl)-phenylalanine
- (2S)-2-[[(Z)-octadec-9-enoyl]amino]-3-phenylpropanoic acid
- CHEMBL4228763
- N-[(9Z)-1-oxo-9-octadecen-1-yl]-L-phenylalanine
- (2S)-2-[(9Z)-octadec-9-enoylamino]-3-phenylpropanoic acid
- (2S)-2-[(9Z)-octadec-9-enoylamino]-3-phenylpropionic acid
- (2S)-2-[(9Z)-octadec-9-enoylamino]-3-phenylpropanoate
- (2S)-2-[(9Z)-octadec-9-enoylamino]-3-phenylpropionate
- N-Oleoyl-Phenylalanine
- SCHEMBL15285227
- CHEBI:134021
- DTXSID201347279
- BDBM50537028
- LMFA08020092
- DA-76245
- HY-138207
- N-[(9Z)-octadec-9-enoyl]-L-phenylalanine
- CS-0145947
- Q63395331
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
429.6 g/mol
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
XLogP3-AA
Property Value
8.6
Reference
Computed by XLogP3 3.0 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Donor Count
Property Value
2
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Acceptor Count
Property Value
3
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Rotatable Bond Count
Property Value
19
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Exact Mass
Property Value
429.32429423 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Monoisotopic Mass
Property Value
429.32429423 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Topological Polar Surface Area
Property Value
66.4 Ų
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Heavy Atom Count
Property Value
31
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
483
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2021.05.07)
Fatty Acyls [FA] -> Fatty amides [FA08] -> N-acyl amino acids [FA0805]
Spectra ID
Ionization Mode
Negative
Top 5 Peaks
164.0721 100
91.0559 61.82
147.0456 19.01
280.2646 17.82
292.2637 4.12
Spectra ID
Ionization Mode
Negative
Top 5 Peaks
428.3169 100
164.0717 2.40
Accession ID
Authors
BGC, Helmholtz Zentrum Muenchen
Instrument
maXis plus UHR-ToF-MS, Bruker Daltonics
Instrument Type
LC-ESI-QTOF
MS Level
MS2
Ionization Mode
POSITIVE
Ionization
ESI
Collision Energy
10
Fragmentation Mode
CID
Column Name
BEH C18 1.7um, 2.1x100mm, Waters
Retention Time
7.193 min
Precursor m/z
430.3316
Precursor Adduct
[M+H]+
Top 5 Peaks
430.332 999
166.0857 97
384.3267 8
412.3221 7
120.0812 5
License
CC BY
Accession ID
Authors
BGC, Helmholtz Zentrum Muenchen
Instrument
maXis plus UHR-ToF-MS, Bruker Daltonics
Instrument Type
LC-ESI-QTOF
MS Level
MS2
Ionization Mode
POSITIVE
Ionization
ESI
Collision Energy
20
Fragmentation Mode
CID
Column Name
BEH C18 1.7um, 2.1x100mm, Waters
Retention Time
7.193 min
Precursor m/z
430.3316
Precursor Adduct
[M+H]+
Top 5 Peaks
166.0855 999
430.3314 139
120.0806 134
167.0887 91
384.3256 46
License
CC BY
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Stereo Count
Same Isotope Count
Same Parent, Connectivity Count
Same Parent, Stereo Count
Same Parent, Isotope Count
Same Parent, Exact Count
Mixtures, Components, and Neutralized Forms Count
Similar Compounds (2D)
Similar Conformers (3D)
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=UWKNPULCJWBBDD-JRUKXMRZSA-N
- ChEBIN-oleoyl-L-phenylalaninehttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:134021
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- EPA DSSToxN-Oleoyl phenylalaninehttps://comptox.epa.gov/dashboard/DTXSID201347279
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingN-Oleoyl phenylalaninehttp://www.hmdb.ca/metabolites/HMDB0062336HMDB0062336_msms_2236574https://hmdb.ca/metabolites/HMDB0062336#spectra
- LIPID MAPSLipid Classificationhttps://www.lipidmaps.org/
- MassBank EuropeN-Oleoyl-Phenylalaninehttps://massbank.eu/MassBank/Result.jsp?inchikey=UWKNPULCJWBBDD-JRUKXMRZSA-N
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics WorkbenchN-oleoyl phenylalaninehttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=4578
- Springer Nature
- WikidataN-oleoyl-L-phenylalaninehttps://www.wikidata.org/wiki/Q63395331
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 396484528https://pubchem.ncbi.nlm.nih.gov/substance/396484528
CONTENTS