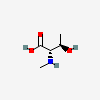
N-Methyl-L-threonine
- 2812-28-4
- N-Methyl-L-threonine
- H-N-Me-Thr-OH
- N-Me-Thr-OH
- methyl-l-threonine
- Create:2006-07-29
- Modify:2025-01-18
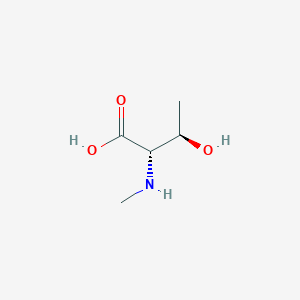

- 2812-28-4
- N-Methyl-L-threonine
- H-N-Me-Thr-OH
- N-Me-Thr-OH
- methyl-l-threonine
- n-methylthreonine
- (2S,3R)-3-Hydroxy-2-(methylamino)butanoic acid
- N-methylidene-L-threonine
- (2S,3R)-3-hydroxy-2-(methylazaniumyl)butanoate
- H-L-MeThr-OH
- N-Methyl-L-threonine; N-Methylthreonine;
- N-alpha-Methyl-L-Threonine
- MFCD02094308
- H-MeThr-OH
- SCHEMBL162004
- DTXSID30426638
- CCAIIPMIAFGKSI-DMTCNVIQSA-N
- AKOS006276574
- CS-W011082
- HY-W010366
- N-Methyl-L-threonine, >=98% (TLC)
- 21148-57-2
- AS-56490
- Q27463992
70.06577 100
88.07634 94.19
116.0711 75.88
134.0714 73.45
98.06038 28.94
88.039 100
86.02348 10.04
132.06555 7.81
80.65544 2.33
51.74053 1.93
Not Classified
Reported as not meeting GHS hazard criteria by 38 of 38 companies. For more detailed information, please visit ECHA C&L website.
Aggregated GHS information provided per 38 reports by companies from 1 notifications to the ECHA C&L Inventory.
Reported as not meeting GHS hazard criteria per 38 of 38 reports by companies. For more detailed information, please visit ECHA C&L website.
There are 0 notifications provided by 0 of 38 reports by companies with hazard statement code(s).
Information may vary between notifications depending on impurities, additives, and other factors. The percentage value in parenthesis indicates the notified classification ratio from companies that provide hazard codes. Only hazard codes with percentage values above 10% are shown.
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=CCAIIPMIAFGKSI-DMTCNVIQSA-N
- EPA DSSToxN-Methyl-L-threoninehttps://comptox.epa.gov/dashboard/DTXSID30426638
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-notice[No public or meaningful name is available]https://echa.europa.eu/substance-information/-/substanceinfo/100.189.694
- Japan Chemical Substance Dictionary (Nikkaji)
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Protein Data Bank in Europe (PDBe)
- SpectraBaseN-METHYL-L-THREONINEhttps://spectrabase.com/spectrum/LOJpfmW4KUd
- Springer Nature
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- WikidataN-methylidene-L-threoninehttps://www.wikidata.org/wiki/Q27463992
- PubChem
- GHS Classification (UNECE)GHS Classification Treehttp://www.unece.org/trans/danger/publi/ghs/ghs_welcome_e.html
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 388416081https://pubchem.ncbi.nlm.nih.gov/substance/388416081