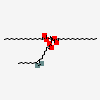
1-Stearoyl-2-stearoyl-3-oleoyl-glycerol
PubChem CID
99649123
Molecular Formula
Synonyms
- TG(18:0/18:0/18:1(9Z))
- 1-stearoyl-2-stearoyl-3-oleoyl-glycerol
- Tracylglycerol(18:0/18:0/18:1n9)
- Tracylglycerol(18:0/18:0/18:1w9)
- TAG(18:0/18:0/18:1n9)
Molecular Weight
889.5 g/mol
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Dates
- Create:2015-12-11
- Modify:2025-01-04
Chemical Structure Depiction
Conformer generation is disallowed since too many atoms, too flexible
[(2S)-2-octadecanoyloxy-3-[(Z)-octadec-9-enoyl]oxypropyl] octadecanoate
Computed by LexiChem 2.6.6 (PubChem release 2019.06.18)
InChI=1S/C57H108O6/c1-4-7-10-13-16-19-22-25-28-31-34-37-40-43-46-49-55(58)61-52-54(63-57(60)51-48-45-42-39-36-33-30-27-24-21-18-15-12-9-6-3)53-62-56(59)50-47-44-41-38-35-32-29-26-23-20-17-14-11-8-5-2/h25,28,54H,4-24,26-27,29-53H2,1-3H3/b28-25-/t54-/m1/s1
Computed by InChI 1.0.5 (PubChem release 2019.06.18)
YFFIQXNTTVSKJC-JRTFMWKKSA-N
Computed by InChI 1.0.5 (PubChem release 2019.06.18)
CCCCCCCCCCCCCCCCCC(=O)OC[C@@H](COC(=O)CCCCCCC/C=C\CCCCCCCC)OC(=O)CCCCCCCCCCCCCCCCC
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C57H108O6
Computed by PubChem 2.1 (PubChem release 2019.06.18)
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
889.5 g/mol
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
XLogP3-AA
Property Value
24.3
Reference
Computed by XLogP3 3.0 (PubChem release 2019.06.18)
Property Name
Hydrogen Bond Donor Count
Property Value
0
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Hydrogen Bond Acceptor Count
Property Value
6
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Rotatable Bond Count
Property Value
55
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Exact Mass
Property Value
888.81459116 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Monoisotopic Mass
Property Value
888.81459116 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Topological Polar Surface Area
Property Value
78.9 Ų
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Heavy Atom Count
Property Value
63
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
978
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2015.09.10)
Solid
329.4 Ų [M+NH4]+ [CCS Type: DT; Buffer gas: N2; Sample Type: Human plasma; Dataset: Ambiguous Lipids]
Lipids -> Ambiguous Lipids
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Stereo Count
Same Isotope Count
Same Parent, Connectivity Count
Same Parent, Stereo Count
Same Parent, Isotope Count
Similar Compounds (2D)
Similar Conformers (3D)
Same Count
- Extracellular
- Membrane
- Baker Lab, Chemistry Department, The University of North Carolina at Chapel HillTG(18:0_18:0_18:1)CCS Classification - Baker Labhttps://tarheels.live/bakerlab/
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutTG(18:0/18:0/18:1(9Z))https://foodb.ca/compounds/FDB098898
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingTG(18:0/18:0/18:1(9Z))http://www.hmdb.ca/metabolites/HMDB0005395
- Japan Chemical Substance Dictionary (Nikkaji)
- Metabolomics WorkbenchTG(18:0/18:0/18:1(9Z))https://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=6543
- PubChem
CONTENTS