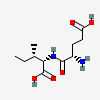
Glutamylisoleucine
- glutamylisoleucine
- Glu-Ile
- L-Glutamyl-L-Isoleucine
- Glutamyl-Isoleucine
- EI dipeptide
- Create:2006-10-25
- Modify:2025-01-18
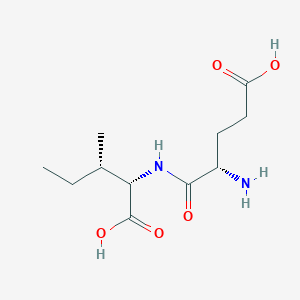

- glutamylisoleucine
- Glu-Ile
- L-Glutamyl-L-Isoleucine
- Glutamyl-Isoleucine
- EI dipeptide
- alpha-Glu-Ile
- E-I Dipeptide
- N-glutamylisoleucine
- alpha-L-Glu-L-Ile
- 5879-22-1
- alpha-glutamylisoleucine
- Glutamate Isoleucine dipeptide
- Glutamate-Isoleucine dipeptide
- alpha-L-glutamyl-L-isoleucine
- L-alpha-glutamyl-L-isoleucine
- SCHEMBL3708036
- glutamic acid isoleucine dipeptide
- CHEBI:141436
- DTXSID801313696
- (2S,3S)-2-[(2S)-2-AMINO-4-CARBOXYBUTANAMIDO]-3-METHYLPENTANOIC ACID
- (4S)-4-amino-5-{[(1S,2S)-1-carboxy-2-methylbutyl]amino}-5-oxopentanoic acid
- E-I
Silke Matysik, Caroline Ivanne Le Roy, Gerhard Liebisch, Sandrine Paule Claus. Metabolomics of fecal samples: A practical consideration. Trends in Food Science & Technology. Vol. 57, Part B, Nov. 2016, p.244-255: http://www.sciencedirect.com/science/article/pii/S0924224416301984
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=SNFUTDLOCQQRQD-ZKWXMUAHSA-N
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/L-Glutamyl-L-isoleucinehttps://commonchemistry.cas.org/detail?cas_rn=5879-22-1
- EPA DSSToxL-Glutamyl-L-isoleucinehttps://comptox.epa.gov/dashboard/DTXSID801313696CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingGlutamylisoleucinehttp://www.hmdb.ca/metabolites/HMDB0028822
- ChEBI
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutGlutamylisoleucinehttps://foodb.ca/compounds/FDB111866
- Metabolomics Workbench
- Springer Nature
- WikidataGlutamylisoleucinehttps://www.wikidata.org/wiki/Q76411589
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 391584896https://pubchem.ncbi.nlm.nih.gov/substance/391584896