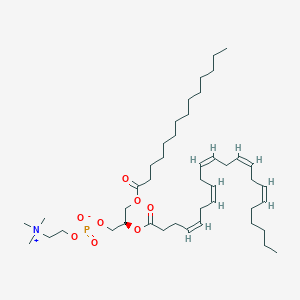
PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))
- PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))
- PC(36:5)
- 1-tetradecanoyl-2-(4Z,7Z,10Z,13Z,16Z-docosapentaenoyl)-sn-glycero-3-phosphocholine
- CHEBI:89583
- GPCho(14:0/22:5n6)
- Create:2011-11-09
- Modify:2025-01-04
- PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))
- PC(36:5)
- 1-tetradecanoyl-2-(4Z,7Z,10Z,13Z,16Z-docosapentaenoyl)-sn-glycero-3-phosphocholine
- CHEBI:89583
- GPCho(14:0/22:5n6)
- GPCho(14:0/22:5w6)
- LMGP01012130
- Phosphatidylcholine(14:0/22:5n6)
- Phosphatidylcholine(14:0/22:5w6)
- PC(14:0/22:5n6)
- PC(14:0/22:5w6)
- 1-myristoyl-2-osbondoyl-sn-glycero-3-phosphocholine
- Q27161780
286.6 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
286.7 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
275 Ų [M-CH3]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
289.3 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
297.3 Ų [M+HCOO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
285.7 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
291.3 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
297.9 Ų [M+HCOO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
291.5 Ų [M+HCOO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
297.8 Ų [M+CH3COO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
288 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
296.7 Ų [M+HCOO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
292.1 Ų [M+HCOO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
286.9 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
284.9 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
289.9 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
285.6 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
289.4 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
275.6 Ų [M-CH3]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
284.8 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
290 Ų [M+H]+ [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
292.7 Ų [M+HCOO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
290.9 Ų [M+HCOO]- [CCS Type: TIMS; Method: calibrated with 4 ions from ESI LC/MS tuning mix (Agilent)]
- Extracellular
- Membrane
The Merck Manual, 17th ed. Mark H. Beers, MD, Robert Berkow, MD, eds. Whitehouse Station, NJ: Merck Research Labs, 1999.
Metabolomics reveals determinants of weight loss during lifestyle intervention in obese children
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- ChEBIPC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:89583
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))https://www.wikidata.org/wiki/Q27161780LOTUS Treehttps://lotus.naturalproducts.net/
- Yeast Metabolome Database (YMDB)PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))https://www.ymdb.ca/compounds/YMDB01824
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingPC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))http://www.hmdb.ca/metabolites/HMDB0007890
- LIPID MAPSLipid Classificationhttps://www.lipidmaps.org/
- Natural Product Activity and Species Source (NPASS)
- MarkerDBLICENSEThis work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.https://markerdb.ca/PC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))https://markerdb.ca/chemicals/2184
- Metabolomics WorkbenchPC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))https://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=14532
- WikidataPC(14:0/22:5(4Z,7Z,10Z,13Z,16Z))https://www.wikidata.org/wiki/Q27161780
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
 CID 53478612 (CID 53478612)
CID 53478612 (CID 53478612)