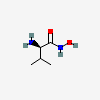
Hydroxyaminovaline
PubChem CID
444599
Molecular Formula
Synonyms
- HYDROXYAMINOVALINE
- 88244-32-0
- DTXSID10332162
- HAV
- (2R)-2-amino-N-hydroxy-3-methylbutanamide
Molecular Weight
132.16 g/mol
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Dates
- Create:2005-06-24
- Modify:2025-01-18
Chemical Structure Depiction
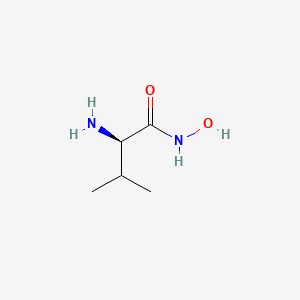
SVG Image

IUPAC Condensed
H-D-Val-NHOH
Sequence
V
IUPAC
D-valine hydroxyamide
(2R)-2-amino-N-hydroxy-3-methylbutanamide
Computed by Lexichem TK 2.7.0 (PubChem release 2021.05.07)
InChI=1S/C5H12N2O2/c1-3(2)4(6)5(8)7-9/h3-4,9H,6H2,1-2H3,(H,7,8)/t4-/m1/s1
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
USSBBYRBOWZYSB-SCSAIBSYSA-N
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
CC(C)[C@H](C(=O)NO)N
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C5H12N2O2
Computed by PubChem 2.1 (PubChem release 2021.05.07)
88244-32-0
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
132.16 g/mol
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
XLogP3-AA
Property Value
-0.5
Reference
Computed by XLogP3 3.0 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Donor Count
Property Value
3
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Acceptor Count
Property Value
3
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Rotatable Bond Count
Property Value
2
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Exact Mass
Property Value
132.089877630 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Monoisotopic Mass
Property Value
132.089877630 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Topological Polar Surface Area
Property Value
75.4 Ų
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Heavy Atom Count
Property Value
9
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
103
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2019.01.04)
Pharmaceuticals -> Listed in ZINC15
S55 | ZINC15PHARMA | Pharmaceuticals from ZINC15 | DOI:10.5281/zenodo.3247749
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Parent, Connectivity Count
Same Parent, Exact Count
Mixtures, Components, and Neutralized Forms Count
Similar Compounds (2D)
Similar Conformers (3D)
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=USSBBYRBOWZYSB-SCSAIBSYSA-N
- ClinicalTrials.govLICENSEThe ClinicalTrials.gov data carry an international copyright outside the United States and its Territories or Possessions. Some ClinicalTrials.gov data may be subject to the copyright of third parties; you should consult these entities for any additional terms of use.https://clinicaltrials.gov/ct2/about-site/terms-conditions#Use
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_useHydroxyaminovalinehttps://www.drugbank.ca/drugs/DB02697
- Therapeutic Target Database (TTD)Hydroxyaminovalinehttps://idrblab.net/ttd/data/drug/details/D05HOS
- EPA DSSToxHYDROXYAMINOVALINEhttps://comptox.epa.gov/dashboard/DTXSID10332162CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Japan Chemical Substance Dictionary (Nikkaji)
- Metabolomics Workbench
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/HydroxyaminovalineNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- WikidataHydroxyaminovalinehttps://www.wikidata.org/wiki/Q27093644
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 390971361https://pubchem.ncbi.nlm.nih.gov/substance/390971361
CONTENTS