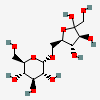
Palatinose
- Palatinose
- Isomaltulose
- 6-O-alpha-D-Glucopyranosyl-D-fructofuranose
- Isomaltulose hydrate
- (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-[[(2R,3S,4S)-3,4,5-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methoxy]oxane-3,4,5-triol
- Create:2004-09-16
- Modify:2025-01-18
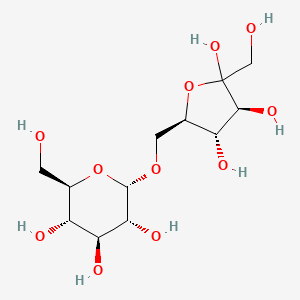

- 6-O alpha-D-glucopyranosyl-D-fructose
- D-fructose, 6-O-alpha-D-glucopyranosyl-, monohydrate
- isomaltulose
- isomaltulose anhydrous
- isomaltulose monohydrate
- palatinose
- palatinose monohydrate
- Palatinose
- Isomaltulose
- 6-O-alpha-D-Glucopyranosyl-D-fructofuranose
- Isomaltulose hydrate
- (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-[[(2R,3S,4S)-3,4,5-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methoxy]oxane-3,4,5-triol
- CHEBI:18394
- 58166-27-1
- (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-(((2R,3S,4S)-3,4,5-trihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)methoxy)tetrahydro-2H-pyran-3,4,5-triol
- (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-[[(2R,3S,4S)-5-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxolan-2-yl]methoxy]oxane-3,4,5-triol
- (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-(((2R,3S,4S)-3,4,5-trihydroxy-5-(hydroxymethyl)oxolan-2-yl)methoxy)oxane-3,4,5-triol
- (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-(((2R,3S,4S)-5-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxolan-2-yl)methoxy)oxane-3,4,5-triol
- SCHEMBL118910
- MFCD00076094
- STL565150
- AKOS037623354
- PD124262
- P1234
- C01742
- J-006975
- Q27103048
- (2R,3S,4S,5R,6S)-2-methylol-6-[[(2R,3S,4S)-3,4,5-trihydroxy-5-methylol-tetrahydrofuran-2-yl]methoxy]tetrahydropyran-3,4,5-triol
177.9 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
176 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
177.6 Ų [M+K]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
177.31 Ų [M+Na]+ [CCS Type: DT; Method: stepped-field]
172.6 Ų [M-H]- [CCS Type: DT; Method: stepped-field]
176.2 Ų [M+Na]+
175.4 Ų [M-H]-
177.7 Ų [M+K]+
103 100
147 96.10
204 75.88
217 69.67
129 54.35
192.9773 999
276.9304 684
278.9421 562
178.9655 410
296.9911 377
101.0234 999
179.0539 848
71.0148 822
89.0237 598
221.0646 583
59.017410 100
89.026062 75.46
89.028717 62.58
59.019581 62.58
71.017311 62.58
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=PVXPPJIGRGXGCY-TZLCEDOOSA-N
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/PalatinoseNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBI6-O-alpha-D-glucopyranosyl-D-fructofuranosehttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:18394
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/Palatinosehttps://www.wikidata.org/wiki/Q27103048LOTUS Treehttps://lotus.naturalproducts.net/
- ClinicalTrials.govLICENSEThe ClinicalTrials.gov data carry an international copyright outside the United States and its Territories or Possessions. Some ClinicalTrials.gov data may be subject to the copyright of third parties; you should consult these entities for any additional terms of use.https://clinicaltrials.gov/ct2/about-site/terms-conditions#Use
- European Chemicals Agency (ECHA)LICENSEUse of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page.https://echa.europa.eu/web/guest/legal-notice6-O-α-D-glucopyranosyl-α-D-fructofuranosehttps://echa.europa.eu/substance-information/-/substanceinfo/100.055.573
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlCompounds with biological roleshttp://www.genome.jp/kegg-bin/get_htext?br08001.keg
- Natural Product Activity and Species Source (NPASS)
- MassBank Europe
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics Workbench
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-lawPalatinosehttp://www.nist.gov/srd/nist1a.cfm
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- Springer Nature
- Wikidata6-O-alpha-D-glucopyranosyl-D-fructofuranosehttps://www.wikidata.org/wiki/Q27103048
- WikipediaIsomaltulosehttps://en.wikipedia.org/wiki/Isomaltulose
- PubChem
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmlisomaltulosehttps://www.ncbi.nlm.nih.gov/mesh/67008189
- Glycan Naming and Subsumption Ontology (GNOme)GNOme
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 389231382https://pubchem.ncbi.nlm.nih.gov/substance/389231382
- NCBI