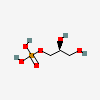
Sn-glycerol-1-phosphate
PubChem CID
439276
Molecular Formula
Synonyms
- sn-glycerol-1-phosphate
- L-Glycerol 1-phosphate
- 5746-57-6
- Glyceryl 1-phosphate, (S)-
- L-alpha-glycerophosphate
Molecular Weight
172.07 g/mol
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Dates
- Create:2004-09-16
- Modify:2025-01-11
Description
Sn-glycerol 1-phosphate is an optically active glycerol 1-phosphate having (S)-configuration. It has a role as an archaeal metabolite. It is a member of sn-glycerol 1-phosphates and a glycerol 1-phosphate. It is a conjugate acid of a sn-glycerol 1-phosphate(2-). It is an enantiomer of a sn-glycerol 3-phosphate.
Sn-Glycerol-1-phosphate is a metabolite found in or produced by Escherichia coli (strain K12, MG1655).
Sn-glycerol-1-phosphate has been reported in Musca domestica and Apis cerana with data available.
Chemical Structure Depiction
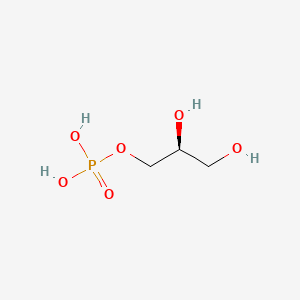
IUPAC
O1-phosphono-sn-glycerol
[(2S)-2,3-dihydroxypropyl] dihydrogen phosphate
Computed by Lexichem TK 2.7.0 (PubChem release 2021.10.14)
InChI=1S/C3H9O6P/c4-1-3(5)2-9-10(6,7)8/h3-5H,1-2H2,(H2,6,7,8)/t3-/m0/s1
Computed by InChI 1.0.6 (PubChem release 2021.10.14)
AWUCVROLDVIAJX-VKHMYHEASA-N
Computed by InChI 1.0.6 (PubChem release 2021.10.14)
C([C@@H](COP(=O)(O)O)O)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C3H9O6P
Computed by PubChem 2.2 (PubChem release 2021.10.14)
- sn-glycerol-1-phosphate
- L-Glycerol 1-phosphate
- 5746-57-6
- Glyceryl 1-phosphate, (S)-
- L-alpha-glycerophosphate
- sn-Glycerol 1-phosphate
- L-alpha-Glycerol phosphate
- L-alpha-Glycerophosphoric acid
- [(2S)-2,3-dihydroxypropyl] dihydrogen phosphate
- UNII-196623779E
- D-(glycerol 3-phosphate)
- L-(glycerol 1-phosphate)
- Glycerophosphoric acid L-alpha-form [MI]
- 1,2,3-Propanetriol, 1-(dihydrogen phosphate), (2S)-
- (2S)-2,3-dihydroxypropyl dihydrogen phosphate
- sn-glycerol 1-(dihydrogen phosphate)
- 196623779E
- sn-Gro-1-P
- 1GP
- 3325-00-6
- 6tim
- L-I+/--Glycerophosphate
- (S)-glyceryl 1-phosphate
- SCHEMBL7341
- CHEBI:16221
- AWUCVROLDVIAJX-VKHMYHEASA-N
- GLYCEROL 1-PHOSPHATE, L-
- DTXSID501315267
- L-.ALPHA.-GLYCEROPHOSPHATE
- L-.ALPHA.-GLYCEROL PHOSPHATE
- L-.ALPHA.-GLYCEROPHOSPHORIC ACID
- PD181512
- C00623
- GLYCEROPHOSPHORIC ACID L-.ALPHA.-FORM [MI]
- Q19970447
- sn-Glycerol 1-phosphate lithium salt, >=95.0% (TLC)
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
172.07 g/mol
Reference
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Property Name
XLogP3-AA
Property Value
-2.9
Reference
Computed by XLogP3 3.0 (PubChem release 2021.10.14)
Property Name
Hydrogen Bond Donor Count
Property Value
4
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Hydrogen Bond Acceptor Count
Property Value
6
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Rotatable Bond Count
Property Value
4
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Exact Mass
Property Value
172.01367500 Da
Reference
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Property Name
Monoisotopic Mass
Property Value
172.01367500 Da
Reference
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Property Name
Topological Polar Surface Area
Property Value
107 Ų
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Heavy Atom Count
Property Value
10
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
129
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2021.10.14)
Pharmaceuticals -> Listed in ZINC15
S55 | ZINC15PHARMA | Pharmaceuticals from ZINC15 | DOI:10.5281/zenodo.3247749
MoNA ID
MS Category
Experimental
MS Type
GC-MS
MS Level
MS1
Instrument
Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies
Instrument Type
GC-EI-TOF
Ionization Mode
positive
Retention Time
842.963 sec
Top 5 Peaks
101 100
299 99.30
147 96.80
103 84.78
357 63.06
License
CC BY-SA
MoNA ID
MS Category
Experimental
MS Type
GC-MS
MS Level
MS1
Instrument
Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies
Instrument Type
GC-EI-TOF
Ionization Mode
positive
Retention Time
460.969 sec
Top 5 Peaks
73 100
299 11.81
103 10.81
147 10.71
101 10.41
License
CC BY-SA
MoNA ID
MS Category
Experimental
MS Type
LC-MS
MS Level
MS2
Precursor Type
[M-H]-
Precursor m/z
171.00587
Instrument
UPLC Q-Tof Premier, Waters
Instrument Type
LC-ESI-QTOF
Ionization
ESI
Ionization Mode
negative
Collision Energy
Ramp 5-60 V
Top 5 Peaks
78.9597 100
171.0059 26.71
96.9699 7.87
License
CC BY-SA
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Isotope Count
Same Parent, Connectivity Count
Same Parent, Isotope Count
Same Parent, Exact Count
Mixtures, Components, and Neutralized Forms Count
Similar Compounds (2D)
Similar Conformers (3D)
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=AWUCVROLDVIAJX-VKHMYHEASA-N
The LOTUS Initiative for Open Natural Products Research: frozen dataset union wikidata (with metadata) | DOI:10.5281/zenodo.5794106
A metabolome atlas of the aging mouse brain. Nat Commun. 2021 Oct 15;12(1):6021. DOI:10.1038/s41467-021-26310-y. PMID:34654818; PMCID:PMC8519999.
The Metabolome Atlas of the Aging Mouse Brain: https://mouse.atlas.metabolomics.us
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/L-α-Glycerophosphatehttps://commonchemistry.cas.org/detail?cas_rn=5746-57-6
- ChemIDplusGlyceryl 1-phosphate, (S)-https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0005746576ChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- EPA DSSToxL-α-Glycerophosphatehttps://comptox.epa.gov/dashboard/DTXSID501315267CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linkingGLYCERYL 1-PHOSPHATE, (S)-https://gsrs.ncats.nih.gov/ginas/app/beta/substances/196623779E
- ChEBISn-glycerol 1-phosphatehttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:16221
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/Sn-glycerol-1-phosphatehttps://www.wikidata.org/wiki/Q19970447LOTUS Treehttps://lotus.naturalproducts.net/
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.html
- West Coast Metabolomics Center-UC Davisrac-Glycerol 3-phosphoate
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics WorkbenchSn-glycerol 1-phosphatehttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=50423
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/Glycerophosphoric AcidNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Springer Nature
- Therapeutic Target Database (TTD)3-Phosphoglycerolhttps://idrblab.net/ttd/data/drug/details/D03MUW
- Wikidata(S)-glyceryl 1-phosphatehttps://www.wikidata.org/wiki/Q19970447
- PubChem
- Glycan Naming and Subsumption Ontology (GNOme)GNOme
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 388566284https://pubchem.ncbi.nlm.nih.gov/substance/388566284
CONTENTS