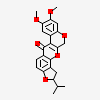
Dehydrodihydrorotenone
- Dehydrodihydrorotenone
- 16,17-dimethoxy-6-propan-2-yl-2,7,20-trioxapentacyclo[11.8.0.03,11.04,8.014,19]henicosa-1(13),3(11),4(8),9,14,16,18-heptaen-12-one
- KBio2_005802
- 16,17-dimethoxy-6-propan-2-yl-2,7,20-trioxapentacyclo(11.8.0.03,11.04,8.014,19)henicosa-1(13),3(11),4(8),9,14,16,18-heptaen-12-one
- Spectrum_000186
- Create:2005-09-12
- Modify:2025-01-11
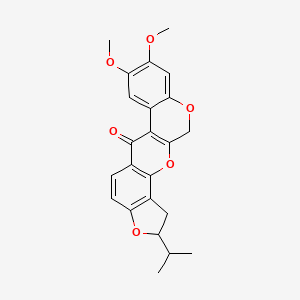
- Dehydrodihydrorotenone
- 16,17-dimethoxy-6-propan-2-yl-2,7,20-trioxapentacyclo[11.8.0.03,11.04,8.014,19]henicosa-1(13),3(11),4(8),9,14,16,18-heptaen-12-one
- KBio2_005802
- 16,17-dimethoxy-6-propan-2-yl-2,7,20-trioxapentacyclo(11.8.0.03,11.04,8.014,19)henicosa-1(13),3(11),4(8),9,14,16,18-heptaen-12-one
- Spectrum_000186
- SpecPlus_000183
- Spectrum2_000184
- Spectrum3_000219
- Spectrum4_001437
- Spectrum5_000294
- BSPBio_001837
- KBioGR_001914
- KBioSS_000666
- DivK1c_006279
- SPBio_000107
- SCHEMBL4278197
- CHEMBL3039066
- CHEBI:93848
- KBio1_001223
- KBio2_000666
- KBio2_003234
- KBio3_001337
- LSM-4404
- CCG-38392
- LMPK12060066
- BRD-A33119430-001-02-2
- BRD-A33119430-001-03-0
- Q27165589
177.94 Ų [M+H-H2O]+ [CCS Type: TW; Method: calibrated with polyalanine and drug standards]
185.29 Ų [M+H]+ [CCS Type: TW; Method: calibrated with polyalanine and drug standards]
279.0619 100
335.1288 76.20
293.0505 68.93
264.0438 61.28
280.0689 49.10
395.1425 100
364.1272 34.39
335.1271 30.55
363.1268 24.73
349.1043 8.68
363.124533 0.30
364.132466 0.25
335.129589 0.22
307.061484 0.06
339.08771 0.04
152.07097365532383 0.14
264.06548594517227 0.08
236.06422473917232 0.05
150.05612865532382 0.05
279.0912154101723 0.05
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- ChEBI
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- LIPID MAPSDehydrodihydrorotenonehttps://lipidmaps.org/databases/lmsd/LMPK12060066Lipid Classificationhttps://www.lipidmaps.org/
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics WorkbenchDehydrodihydrorotenonehttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=22712
- SpectraBase21,22-Dihydro-6,12a-didehydro-rotenonehttps://spectrabase.com/spectrum/LcAMn3vNimo
- Wikidata
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/