3-Oxotetradecanoyl-CoA
PubChem CID
11966197
Molecular Formula
Synonyms
- 3-Oxotetradecanoyl-CoA
- 3-oxomyristoyl-CoA
- 122364-86-7
- [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-[[hydroxy-[hydroxy-[(3R)-3-hydroxy-2,2-dimethyl-3-[
- 3'-phosphoadenosine 5'-{3-[(3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(3-oxotetradecanoyl)sulfanyl]ethyl}amino)propyl]amino}butyl] dihydrogen diphosphate}
Molecular Weight
991.9 g/mol
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Dates
- Create:2006-12-22
- Modify:2025-01-18
Description
3-oxotetradecanoyl-CoA is a 3-oxo-fatty acyl-CoA. It has a role as a human metabolite, a Saccharomyces cerevisiae metabolite, an Escherichia coli metabolite and a mouse metabolite. It is functionally related to a myristoyl-CoA and a 3-oxotetradecanoic acid. It is a conjugate acid of a 3-oxotetradecanoyl-CoA(4-).
3-Oxotetradecanoyl-CoA is a metabolite found in or produced by Escherichia coli (strain K12, MG1655).
3-Oxotetradecanoyl-CoA has been reported in Homo sapiens and Bos taurus with data available.
Chemical Structure Depiction
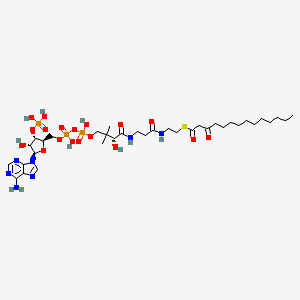
Conformer generation is disallowed since too many atoms, too flexible
S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-hydroxyphosphoryl]oxy-2-hydroxy-3,3-dimethylbutanoyl]amino]propanoylamino]ethyl] 3-oxotetradecanethioate
Computed by Lexichem TK 2.7.0 (PubChem release 2021.05.07)
InChI=1S/C35H60N7O18P3S/c1-4-5-6-7-8-9-10-11-12-13-23(43)18-26(45)64-17-16-37-25(44)14-15-38-33(48)30(47)35(2,3)20-57-63(54,55)60-62(52,53)56-19-24-29(59-61(49,50)51)28(46)34(58-24)42-22-41-27-31(36)39-21-40-32(27)42/h21-22,24,28-30,34,46-47H,4-20H2,1-3H3,(H,37,44)(H,38,48)(H,52,53)(H,54,55)(H2,36,39,40)(H2,49,50,51)/t24-,28-,29-,30+,34-/m1/s1
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
IQNFBGHLIVBNOU-QSGBVPJFSA-N
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
CCCCCCCCCCCC(=O)CC(=O)SCCNC(=O)CCNC(=O)[C@@H](C(C)(C)COP(=O)(O)OP(=O)(O)OC[C@@H]1[C@H]([C@H]([C@@H](O1)N2C=NC3=C(N=CN=C32)N)O)OP(=O)(O)O)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C35H60N7O18P3S
Computed by PubChem 2.1 (PubChem release 2021.05.07)
- 3-Oxotetradecanoyl-CoA
- 3-oxomyristoyl-CoA
- 122364-86-7
- [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-[[hydroxy-[hydroxy-[(3R)-3-hydroxy-2,2-dimethyl-3-[
- 3'-phosphoadenosine 5'-{3-[(3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(3-oxotetradecanoyl)sulfanyl]ethyl}amino)propyl]amino}butyl] dihydrogen diphosphate}
- S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-hydroxyphosphoryl]oxy-2-hydroxy-3,3-dimethylbutanoyl]amino]propanoylamino]ethyl] 3-oxotetradecanethioate
- CHEBI:28726
- S-(3-oxotetradecanoate
- 3-ketotetradecanoyl-CoA
- 3-oxomyristoyl-Coenzyme A
- S-(3-oxotetradecanoic acid
- S-(3-oxotetradecanoate) CoA
- SCHEMBL1332729
- DTXSID101294130
- S-(3-oxotetradecanoate) Coenzyme A
- LMFA07050256
- Coenzyme A, S-(3-oxotetradecanoate)
- 3-oxomyristoyl-CoA; (Acyl-CoA); [M+H]+;
- C05261
- Q27103864
- 5'-{3-[(3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(3-oxotetradecanoyl)sulfanyl]ethyl}amino)propyl]amino}butyl] dihydrogen diphosphate}3'-phosphoadenosine
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
991.9 g/mol
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
XLogP3-AA
Property Value
-0.6
Reference
Computed by XLogP3 3.0 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Donor Count
Property Value
9
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Acceptor Count
Property Value
23
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Rotatable Bond Count
Property Value
32
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Exact Mass
Property Value
991.29284026 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Monoisotopic Mass
Property Value
991.29284026 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Topological Polar Surface Area
Property Value
406 Ų
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Heavy Atom Count
Property Value
64
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
1670
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
5
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2019.01.04)
Solid
Fatty Acyls [FA] -> Fatty esters [FA07] -> Fatty acyl CoAs [FA0705]
MoNA ID
MS Category
Experimental
MS Type
Other
Precursor Type
[M+H]+
Precursor m/z
992.30012
Top 5 Peaks
485.30438 100
428.03724 45.05
992.30012 40.04
136.06232 5.01
583.28127 5.01
MoNA ID
MS Category
Experimental
MS Type
Other
Precursor Type
[M+H]+
Precursor m/z
992.30012
Top 5 Peaks
485.30438 100
428.03724 45.05
992.30012 40.04
136.06232 5.01
583.28127 5.01
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Parent, Connectivity Count
Same Parent, Exact Count
Mixtures, Components, and Neutralized Forms Count
Similar Compounds (2D)
Similar Conformers (3D)
- Extracellular
- Membrane
- Carnitine palmitoyl transferase deficiency (I)
- Carnitine palmitoyl transferase deficiency (II)
- Ethylmalonic Encephalopathy
- Fatty Acid Elongation In Mitochondria
- Fatty acid Metabolism
- Glutaric Aciduria Type I
- Long chain acyl-CoA dehydrogenase deficiency (LCAD)
- Long-chain-3-hydroxyacyl-coa dehydrogenase deficiency (LCHAD)
- Medium chain acyl-coa dehydrogenase deficiency (MCAD)
- Mitochondrial Beta-Oxidation of Long Chain Saturated Fatty Acids
- Total 13 pathways, visit the HMDB page for details
WormJam Metabolites Local CSV for MetFrag | DOI:10.5281/zenodo.3403364
WormJam: A consensus C. elegans Metabolic Reconstruction and Metabolomics Community and Workshop Series, Worm, 6:2, e1373939, DOI:10.1080/21624054.2017.1373939
Zebrafish Pathway Metabolite MetFrag Local CSV (Beta) | DOI:10.5281/zenodo.3457553
The LOTUS Initiative for Open Natural Products Research: frozen dataset union wikidata (with metadata) | DOI:10.5281/zenodo.5794106
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/Coenzyme A, S-(3-oxotetradecanoate)https://commonchemistry.cas.org/detail?cas_rn=122364-86-7
- EPA DSSToxCoenzyme A, S-(3-oxotetradecanoate)https://comptox.epa.gov/dashboard/DTXSID101294130CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citing3-Oxotetradecanoyl-CoAhttp://www.hmdb.ca/metabolites/HMDB0003935
- ChEBI3-oxotetradecanoyl-CoAhttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:28726
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/3-Oxotetradecanoyl-CoAhttps://www.wikidata.org/wiki/Q27103864LOTUS Treehttps://lotus.naturalproducts.net/
- Yeast Metabolome Database (YMDB)3-oxotetradecanoyl-CoAhttps://www.ymdb.ca/compounds/YMDB00478
- ECI Group, LCSB, University of Luxembourg3-oxotetradecanoyl-CoA
- Natural Product Activity and Species Source (NPASS)
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/about3-Oxotetradecanoyl-CoAhttps://foodb.ca/compounds/FDB023254
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.html
- LIPID MAPSLipid Classificationhttps://www.lipidmaps.org/
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics Workbench3-oxotetradecanoyl-CoAhttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=51827
- Wikidata3-oxotetradecanoyl-CoAhttps://www.wikidata.org/wiki/Q27103864
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
CONTENTS