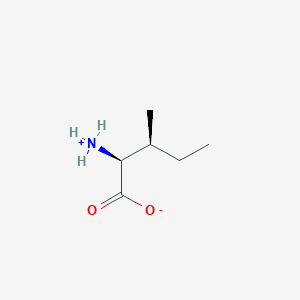
(2S,3S)-2-ammonio-3-methylpentanoate
- (2S,3S)-2-ammonio-3-methylpentanoate
- (2S,3S)-2-azaniumyl-3-methylpentanoate
- L-isoleucine zwitterion
- 3f6g
- 3f6h
- Create:2006-07-29
- Modify:2025-01-18

158.15 100
218.1 20.02
159.2 14.14
147.15 10.81
100.1 8.88
158.0 100
218.0 47.65
147.0 31.63
100.0 29.13
232.0 13.31
86.09078 100
69.06478 96.70
44.04484 52.60
41.03405 21.90
57.05235 16.10
86.09962 100
69.07308 6.10
44.05154 4.30
- All Tissues
- Placenta
- Prostate
- Cytoplasm
- Extracellular
- Mitochondria
- 2-Methyl-3-Hydroxybutryl CoA Dehydrogenase Deficiency
- 3-Hydroxy-3-Methylglutaryl-CoA Lyase Deficiency
- 3-hydroxyisobutyric acid dehydrogenase deficiency
- 3-hydroxyisobutyric aciduria
- 3-Methylcrotonyl Coa Carboxylase Deficiency Type I
- 3-Methylglutaconic Aciduria Type I
- 3-Methylglutaconic Aciduria Type III
- 3-Methylglutaconic Aciduria Type IV
- Amikacin Action Pathway
- Arbekacin Action Pathway
- Total 48 pathways, visit the HMDB page for details
PubMed: 12101068, 10508118, 10472531, 19551947, 11978597, 10234605, 6422161, 23430924, 18088602
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
Peritoneal dialysis in maple-syrup-urine disease: Studies on branched-chain amino and keto acids. Eur J Pediatr (1980) 134: 57. https://doi.org/10.1007/BF00442404
Silke Matysik, Caroline Ivanne Le Roy, Gerhard Liebisch, Sandrine Paule Claus. Metabolomics of fecal samples: A practical consideration. Trends in Food Science & Technology. Vol. 57, Part B, Nov. 2016, p.244-255: http://www.sciencedirect.com/science/article/pii/S0924224416301984
The Merck Manual, 17th ed. Mark H. Beers, MD, Robert Berkow, MD, eds. Whitehouse Station, NJ: Merck Research Labs, 1999.
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/L-Isoleucine polymerhttps://commonchemistry.cas.org/detail?cas_rn=34464-35-2
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingL-Isoleucinehttp://www.hmdb.ca/metabolites/HMDB0000172HMDB0000172_cms_30031https://hmdb.ca/metabolites/HMDB0000172#spectra
- ChEBIL-isoleucine zwitterionhttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:58045
- Yeast Metabolome Database (YMDB)L-Isoleucinehttps://www.ymdb.ca/compounds/YMDB00038
- Crystallography Open Database (COD)LICENSEAll data in the COD and the database itself are dedicated to the public domain and licensed under the CC0 License. Users of the data should acknowledge the original authors of the structural data.https://creativecommons.org/publicdomain/zero/1.0/
- The Cambridge Structural Database
- SpectraBaseL-isoleucinehttps://spectrabase.com/spectrum/Jf4QBaCcSYL-Isoleucinehttps://spectrabase.com/spectrum/EdZxSpzEdo0L-ISOLEUCINEhttps://spectrabase.com/spectrum/62k29n4r6cKL-Isoleucinehttps://spectrabase.com/spectrum/CEYzMeGJ13lL-ISOLEUCINEhttps://spectrabase.com/spectrum/1ydI9fNPbJ9L-Isoleucinehttps://spectrabase.com/spectrum/8iUxmBFEpgfL-Isoleucinehttps://spectrabase.com/spectrum/BVCHVWbfWxZL-Isoleucinehttps://spectrabase.com/spectrum/LxKPtOIp9fhL-Isoleucinehttps://spectrabase.com/spectrum/KeGmMPX45qAL-Isoleucinehttps://spectrabase.com/spectrum/FWK2f5U0QEm
- LIPID MAPS2S-Amino-3S-methylpentanoic acidhttps://lipidmaps.org/databases/lmsd/LMFA01100047Lipid Classificationhttps://www.lipidmaps.org/
- Natural Product Activity and Species Source (NPASS)
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- WikidataL-isoleucine zwitterionhttps://www.wikidata.org/wiki/Q106345669L-isoleucinehttps://www.wikidata.org/wiki/Q484940
- PubChem
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/LOTUS Treehttps://lotus.naturalproducts.net/

 CID 6306 (l-Isoleucine)
CID 6306 (l-Isoleucine)







