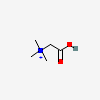
N,N,N-trimethylglycinium
- TRIMETHYL GLYCINE
- N,N,N-trimethylglycinium
- Cystadane
- carboxymethyl(trimethyl)azanium
- carboxy-N,N,N-trimethylmethanaminium
- Create:2004-09-16
- Modify:2024-12-27
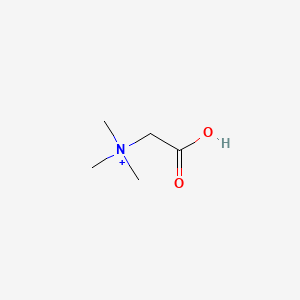
- Acidin Pepsin
- Acidin-Pepsin
- AcidinPepsin
- Betaine
- Betaine Hydrochloride
- Betaine, Glycine
- C.B.B.
- Citrate de Bétaïne Beaufour
- Citrate de Bétaïne UPSA
- Cystadane
- Glycine Betaine
- Hepastyl
- Hydrochloride, Betaine
- Lycine
- Novobetaine
- Oxyneurine
- Scorbo bétaïne
- Scorbo-bétaïne
- Scorbobétaïne
- Stea 16
- Stea-16
- Stea16
- TRIMETHYL GLYCINE
- N,N,N-trimethylglycinium
- Cystadane
- carboxymethyl(trimethyl)azanium
- carboxy-N,N,N-trimethylmethanaminium
- Loramine AMB-13
- CHEBI:41139
- 6640-00-2
- Aminocoat
- Betafin
- Greenstim
- FinnStim
- Betafin BP
- Betafin BCR
- .alpha.-Earleine
- (Carboxymethyl)trimethylammonium hydroxide inner salt
- FEMA NO. 4223
- NCGC00015150-03
- NCGC00015150-04
- BET
- 1-carboxy-n,n,n-trimethyl-methanaminium
- a-Earleine
- Acidin Pepsin
- SR-01000075714
- Spectrum_001264
- trimethylbetaine Glycine
- 1r9l
- 1sw2
- 2b4l
- Spectrum2_000880
- Spectrum4_000735
- Spectrum5_001577
- Lopac-B-3501
- (Trimethylammonio)acetate #
- NCIOpen2_001196
- Lopac0_000172
- BSPBio_002550
- KBioGR_001030
- KBioSS_001744
- AMMONIUM (CARBOXYMETHYL)-TRIMETHYL-PALMITATE
- (carboxymethyl)trimethylazanium
- CHEMBL95889
- DivK1c_000642
- SCHEMBL134333
- SPBio_000959
- KBio1_000642
- KBio2_001744
- KBio2_004312
- KBio2_006880
- DTXSID40859190
- KWIUHFFTVRNATP-UHFFFAOYSA-O
- NINDS_000642
- NSC12179
- NSC87835
- BDBM50357226
- NSC-12179
- NSC-87835
- CCG-204267
- DB04455
- Betaine, BioUltra, >=99.0% (NT)
- IDI1_000642
- NCGC00015150-01
- NCGC00015150-02
- NCGC00162080-01
- SBI-0050160.P003
- Betaine, >=98% (perchloric acid titration)
- NS00068580
- S3755
- 1-carboxy-N,N,N-trimethyl-Methanaminium hydroxide
- Betaine, certified reference material, TraceCERT(R)
- SR-01000075714-8
- BRD-K12014493-001-02-2
- Q27095239
- Betaine, United States Pharmacopeia (USP) Reference Standard
- 6915-17-9
133.8 Ų [M+Na]+
121.1 Ų [M+H]+
130.4 Ų [M+K]+
118.024 100
59.049 7.61
58.26 100
59.337 50.77
117.939 8.65
118.086 999
119.0894 76
118.0862 999
119.0896 81
59.072678 0.59
58.06487 0.41
118.0861 100
59.0728 4
58.0650 3.70
- Adipose Tissue
- Adrenal Cortex
- Adrenal Gland
- Bladder
- Epidermis
- Fibroblasts
- Intestine
- Kidney
- Liver
- Ovary
- Pancreas
- Placenta
- Platelet
- Prostate
- Skeletal Muscle
- Spleen
- Testis
- Cytoplasm
- Extracellular
- Mitochondria
- 3-Phosphoglycerate dehydrogenase deficiency
- Cystathionine Beta-Synthase Deficiency
- Dihydropyrimidine Dehydrogenase Deficiency (DHPD)
- Dimethylglycine Dehydrogenase Deficiency
- Dimethylglycine Dehydrogenase Deficiency
- Glycine N-methyltransferase Deficiency
- Glycine, serine and threonine metabolism
- Glycine, serine and threonine metabolism
- Homocystinuria-megaloblastic anemia due to defect in cobalamin metabolism, cblG complementation type
- Hyperglycinemia, non-ketotic
- Total 18 pathways, visit the HMDB page for details
Silke Matysik, Caroline Ivanne Le Roy, Gerhard Liebisch, Sandrine Paule Claus. Metabolomics of fecal samples: A practical consideration. Trends in Food Science & Technology. Vol. 57, Part B, Nov. 2016, p.244-255: http://www.sciencedirect.com/science/article/pii/S0924224416301984
The Merck Manual, 17th ed. Mark H. Beers, MD, Robert Berkow, MD, eds. Whitehouse Station, NJ: Merck Research Labs, 1999.
PubMed: 5075233, 19551947, 12408190
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
PubMed: 19809936, 19551947, 2226555, 28853722
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
PubMed: 12101068, 10508118, 10472531, 19551947, 11978597, 10234605, 6422161, 23430924, 18088602
MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de)
Peritoneal dialysis in maple-syrup-urine disease: Studies on branched-chain amino and keto acids. Eur J Pediatr (1980) 134: 57. https://doi.org/10.1007/BF00442404
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=KWIUHFFTVRNATP-UHFFFAOYSA-O
- ChEBIN,N,N-trimethylglyciniumhttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:41139
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_useN,N,N-trimethylglyciniumhttps://www.drugbank.ca/drugs/DB04455
- Open TargetsLICENSEDatasets generated by the Open Targets Platform are freely available for download.https://platform-docs.opentargets.org/licence
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- ClinicalTrials.govLICENSEThe ClinicalTrials.gov data carry an international copyright outside the United States and its Territories or Possessions. Some ClinicalTrials.gov data may be subject to the copyright of third parties; you should consult these entities for any additional terms of use.https://clinicaltrials.gov/ct2/about-site/terms-conditions#Use
- Crystallography Open Database (COD)LICENSEAll data in the COD and the database itself are dedicated to the public domain and licensed under the CC0 License. Users of the data should acknowledge the original authors of the structural data.https://creativecommons.org/publicdomain/zero/1.0/
- Drugs@FDALICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linking
- DTP/NCILICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuse
- EPA DSSToxCarboxy-N,N,N-trimethylmethanaminiumhttps://comptox.epa.gov/dashboard/DTXSID40859190CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingHMDB0000043_nmr_one_166377https://hmdb.ca/metabolites/HMDB0000043#spectra
- EU Clinical Trials Register
- SpectraBaseBetaine cationhttps://spectrabase.com/spectrum/7p93E8lSxlmBetaine cationhttps://spectrabase.com/spectrum/JNcr626PgET
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-law
- Japan Chemical Substance Dictionary (Nikkaji)
- MarkerDBLICENSEThis work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.https://markerdb.ca/
- MassBank Europe
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics WorkbenchN,N,N-trimethylglyciniumhttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=148901
- Natural Product Activity and Species Source (NPASS)
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/BETAINENORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- WikidataTrimethyl Glycinehttps://www.wikidata.org/wiki/Q27095239
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmlGastrointestinal Agentshttps://www.ncbi.nlm.nih.gov/mesh/68005765Lipotropic Agentshttps://www.ncbi.nlm.nih.gov/mesh/68008082
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 389811382https://pubchem.ncbi.nlm.nih.gov/substance/389811382
- NCBI