2-Monoolein
- 2-Monoolein
- 2-Oleoylglycerol
- 3443-84-3
- 2-Monooleoylglycerol
- 2-oleoyl-glycerol
- Create:2005-03-27
- Modify:2025-01-10
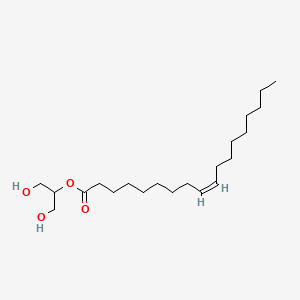
- 2-OG lipid
- 2-oleoylglycerol
- 2-oleyl glycerol ether
- 2-Monoolein
- 2-Oleoylglycerol
- 3443-84-3
- 2-Monooleoylglycerol
- 2-oleoyl-glycerol
- 2-Glyceryl monooleate
- Glyceryl 2-oleate
- Glycerol 2-monooleate
- 2-OG
- 1,3-dihydroxypropan-2-yl oleate
- sn-2-monoolein
- .beta.-Monoolein
- 1,3-dihydroxypropan-2-yl (Z)-octadec-9-enoate
- UNII-9A2389K694
- 2-oleoyl glycerol
- 1,3-dihydroxypropan-2-yl (9Z)-octadec-9-enoate
- 2-mono-C18:1
- MG(0:0/18:1(9Z)/0:0)
- CHEBI:73990
- 2-(9Z-octadecenoyl)-sn-glycerol
- 9A2389K694
- Olein, 2-mono-
- 2-oleyl glycerol ether
- 2-Oleoyl glycerol ether
- 2-oleoylglycerol (2OG)
- DSSTox_CID_7875
- .beta.-Glyceryl monooloeate
- DSSTox_RID_78598
- DSSTox_GSID_27875
- SCHEMBL16172
- 2-(9Z)-octadecenoylglycerol
- 2-Oleoylmonoglycerol (2-OG)
- 2-(9Z-octadecenoyl)-glycerol
- GTPL5112
- 1,3-dihydroxypropan-2-yloleate
- CHEMBL3182200
- DTXSID8058661
- 2-Oleoyl Glycerol (80per cent)
- 2-(9Z-octadecenoyl)-rac-glycerol
- BDBM197165
- 2-Oleoylglycerol, >=94% (TLC)
- Tox21_303545
- 9-Octadecenoic acid (Z)-, 2-hydroxy-1-(hydroxymethyl)ethyl ester
- LMGL01010024
- MFCD00171536
- AKOS027320617
- MAG(0:0/18:1n9)
- MAG(0:0/18:1w9)
- CS-W011837
- HY-W011121
- MG(0:0/18:1n9)
- MG(0:0/18:1w9)
- NCGC00257408-01
- AS-77989
- PD018141
- CAS-25496-72-4
- NS00051170
- F85613
- Q2813830
- BRD-K77087341-001-02-4
- 2-Hydroxy-1-(hydroxymethyl)ethyl (9Z)-9-octadecenoate
- 9-OCTADECENOIC ACID (9Z)-, 2-HYDROXY-1-(HYDROXYMETHYL)ETHYL ESTER
- 2OG
- YOG
103.0 100
129.0 81.48
147.0 24.92
101.0 12.01
218.0 7.91
103 100
129 81.48
147 24.92
101 12.01
218 7.91
- Extracellular
- Membrane
Silke Matysik, Caroline Ivanne Le Roy, Gerhard Liebisch, Sandrine Paule Claus. Metabolomics of fecal samples: A practical consideration. Trends in Food Science & Technology. Vol. 57, Part B, Nov. 2016, p.244-255: http://www.sciencedirect.com/science/article/pii/S0924224416301984
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=UPWGQKDVAURUGE-KTKRTIGZSA-N
- BindingDBLICENSEAll data curated by BindingDB staff are provided under the Creative Commons Attribution 3.0 License (https://creativecommons.org/licenses/by/3.0/us/).https://www.bindingdb.org/rwd/bind/info.jsp2-Oleoylmonoglycerol (2-OG)https://www.bindingdb.org/rwd/bind/chemsearch/marvin/MolStructure.jsp?monomerid=197165
- Comparative Toxicogenomics Database (CTD)LICENSEIt is to be used only for research and educational purposes. Any reproduction or use for commercial purpose is prohibited without the prior express written permission of NC State University.http://ctdbase.org/about/legal.jsp2-oleoylglycerolhttps://ctdbase.org/detail.go?type=chem&acc=C505247
- IUPHAR/BPS Guide to PHARMACOLOGYLICENSEThe Guide to PHARMACOLOGY database is licensed under the Open Data Commons Open Database License (ODbL) https://opendatacommons.org/licenses/odbl/. Its contents are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (http://creativecommons.org/licenses/by-sa/4.0/)https://www.guidetopharmacology.org/about.jsp#licenseGuide to Pharmacology Target Classificationhttps://www.guidetopharmacology.org/targets.jsp
- Therapeutic Target Database (TTD)2-oleoyl glycerolhttps://idrblab.net/ttd/data/drug/details/D05FOV
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/2-Oleoylglycerolhttps://commonchemistry.cas.org/detail?cas_rn=3443-84-3
- ChemIDplus2-Glyceryl monooleatehttps://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0003443843ChemIDplus Chemical Information Classificationhttps://pubchem.ncbi.nlm.nih.gov/source/ChemIDplus
- EPA DSSTox2-Oleoylglycerolhttps://comptox.epa.gov/dashboard/DTXSID8058661CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- FDA Global Substance Registration System (GSRS)LICENSEUnless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required.https://www.fda.gov/about-fda/about-website/website-policies#linkingGLYCERYL 2-OLEATEhttps://gsrs.ncats.nih.gov/ginas/app/beta/substances/9A2389K694
- ChEBI2-oleoylglycerolhttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:73990
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/2-Monooleinhttps://www.wikidata.org/wiki/Q2813830LOTUS Treehttps://lotus.naturalproducts.net/
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/aboutGlyceryl monooleatehttps://foodb.ca/compounds/FDB009514
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingMG(0:0/18:1(9Z)/0:0)http://www.hmdb.ca/metabolites/HMDB0011537HMDB0011537_cms_32368https://hmdb.ca/metabolites/HMDB0011537#spectra
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- NIST Mass Spectrometry Data CenterLICENSEData covered by the Standard Reference Data Act of 1968 as amended.https://www.nist.gov/srd/public-law9-Octadecenoic acid (Z)-, 2-hydroxy-1-(hydroxymethyl)ethyl esterhttp://www.nist.gov/srd/nist1a.cfm
- SpectraBase9-Octadecenoic acid (Z)-, 2-hydroxy-1-(hydroxymethyl)ethyl esterhttps://spectrabase.com/spectrum/Dx7DBdBXl969-Octadecenoic acid (Z)-, 2-hydroxy-1-(hydroxymethyl)ethyl esterhttps://spectrabase.com/spectrum/GjvOPVpplbM
- KNApSAcK Species-Metabolite Database
- Natural Product Activity and Species Source (NPASS)1,3-Dihydroxypropan-2-Yl (Z)-Octadec-9-Enoatehttps://bidd.group/NPASS/compound.php?compoundID=NPC148192
- West Coast Metabolomics Center-UC Davis2-Monoolein
- LIPID MAPSLipid Classificationhttps://www.lipidmaps.org/
- Metabolomics WorkbenchMG(0:0/18:1(9Z)/0:0)https://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=5774
- Nature Chemical Biology
- PharosLICENSEData accessed from Pharos and TCRD is publicly available from the primary sources listed above. Please respect their individual licenses regarding proper use and redistribution.https://pharos.nih.gov/about2-oleoyl glycerolhttps://pharos.nih.gov/ligands/FNNULWSBKQD4
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- Springer Nature
- Wikidataglyceryl 2-oleatehttps://www.wikidata.org/wiki/Q2813830
- Wikipedia2-Oleoylglycerolhttps://en.wikipedia.org/wiki/2-Oleoylglycerol
- PubChem
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.html2-oleoylglycerolhttps://www.ncbi.nlm.nih.gov/mesh/67505247
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/NORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 389000627https://pubchem.ncbi.nlm.nih.gov/substance/389000627


