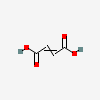
2-Butenedioic acid
PubChem CID
723
Molecular Formula
Synonyms
- 2-Butenedioic acid
- Butenedioic acid
- 2-aButenedioic acid
- 1,2-Ethenedicarboxylic
- NCIOpen2_003792
Molecular Weight
116.07 g/mol
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Dates
- Create:2005-03-26
- Modify:2025-01-18
Description
2-Butenedioic acid has been reported in Phomopsis velata, Inula grandis, and other organisms with data available.
Chemical Structure Depiction
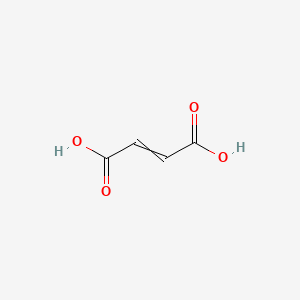
but-2-enedioic acid
Computed by Lexichem TK 2.7.0 (PubChem release 2021.10.14)
InChI=1S/C4H4O4/c5-3(6)1-2-4(7)8/h1-2H,(H,5,6)(H,7,8)
Computed by InChI 1.0.6 (PubChem release 2021.10.14)
VZCYOOQTPOCHFL-UHFFFAOYSA-N
Computed by InChI 1.0.6 (PubChem release 2021.10.14)
C(=CC(=O)O)C(=O)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C4H4O4
Computed by PubChem 2.2 (PubChem release 2021.10.14)
6915-18-0
- (E)-2-butenedioic acid
- 2-butenedioic acid
- ammonium fumarate
- fumarate dianion
- fumarate(2-)
- fumaric acid
- Furamag
- mafusol
- magnesium fumarate
- sodium fumarate
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
116.07 g/mol
Reference
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Property Name
XLogP3
Property Value
-0.3
Reference
Computed by XLogP3 3.0 (PubChem release 2021.10.14)
Property Name
Hydrogen Bond Donor Count
Property Value
2
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Hydrogen Bond Acceptor Count
Property Value
4
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Rotatable Bond Count
Property Value
2
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Exact Mass
Property Value
116.01095860 Da
Reference
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Property Name
Monoisotopic Mass
Property Value
116.01095860 Da
Reference
Computed by PubChem 2.2 (PubChem release 2021.10.14)
Property Name
Topological Polar Surface Area
Property Value
74.6 Ų
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Heavy Atom Count
Property Value
8
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
119
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.10.14)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2021.10.14)
- Boiling point
- Chemical shift
- Corrosion
- Crystal structure
- Density
- Diamagnetic susceptibility
- Formula unit
- Fusion temperature
- Heat of sublimation
- Magnetic anisotropy
- Magnetic susceptibility
- Melting temperature
- Molar conductivity
- Nuclear quadrupole resonance spectroscopy
- Phase transition
- Quadrupole coupling
- Space group
- Surface tension
- Transition enthalpy
- Unit cell
- Unit cell parameter
- Vapor pressure
- Viscosity
Spectra ID
Ionization Mode
Negative
Top 5 Peaks
71.01202 100
114.99653 23.26
73.02469 20.93
Accession ID
Authors
Tetsuya Mori, Center for Sustainable Resource Science, RIKEN
Instrument
LC, Waters Acquity UPLC System; MS, Waters Xevo G2 Q-Tof
Instrument Type
LC-ESI-QTOF
MS Level
MS2
Ionization Mode
NEGATIVE
Ionization
ESI
Collision Energy
6V
Column Name
Acquity bridged ethyl hybrid C18 (1.7 um, 2.1 mm * 100 mm, Waters)
Retention Time
1.66
Precursor m/z
115.0023
Precursor Adduct
[M-H]-
Top 5 Peaks
71.01202 999
114.99653 232
73.02469 209
115.00888 209
License
CC BY-NC-SA
Reference
Tsugawa H., Nakabayashi R., Mori T., Yamada Y., Takahashi M., Rai A., Sugiyama R., Yamamoto H., Nakaya T., Yamazaki M., Kooke R., Bac-Molenaar JA., Oztolan-Erol N., Keurentjes JJB., Arita M., Saito K. (2019) "A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms" Nature Methods 16(4):295-298. [doi:10.1038/s41592-019-0358-2]
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Stereo Count
Same Isotope Count
Same Parent, Connectivity Count
Same Parent, Stereo Count
Same Parent, Isotope Count
Same Parent, Exact Count
Mixtures, Components, and Neutralized Forms Count
Similar Compounds (2D)
Similar Conformers (3D)
PubMed Count
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=VZCYOOQTPOCHFL-UHFFFAOYSA-N
The LOTUS Initiative for Open Natural Products Research: frozen dataset union wikidata (with metadata) | DOI:10.5281/zenodo.5794106
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/2-Butenedioic acidhttps://www.wikidata.org/wiki/Q27109609LOTUS Treehttps://lotus.naturalproducts.net/
- EPA DSSTox2-Butenedioic acidhttps://comptox.epa.gov/dashboard/DTXSID8048157CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingBut-2-enedioic acidhttp://www.hmdb.ca/metabolites/HMDB0249450HMDB0249450_msms_2235714https://hmdb.ca/metabolites/HMDB0249450#spectra
- MassBank EuropeFumaric acid (not validated)https://massbank.eu/MassBank/Result.jsp?inchikey=VZCYOOQTPOCHFL-UHFFFAOYSA-N
- SpectraBaseMaleic acidhttps://spectrabase.com/spectrum/FbJepM5Uve0Fumaric acidhttps://spectrabase.com/spectrum/Ap3gFkGBAne
- Springer Nature
- SpringerMaterials
- Wikidatabutenedioic acidhttps://www.wikidata.org/wiki/Q27109609
- PubChem
- Medical Subject Headings (MeSH)LICENSEWorks produced by the U.S. government are not subject to copyright protection in the United States. Any such works found on National Library of Medicine (NLM) Web sites may be freely used or reproduced without permission in the U.S.https://www.nlm.nih.gov/copyright.htmlfumaric acidhttps://www.ncbi.nlm.nih.gov/mesh/67032005
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/NORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403459920https://pubchem.ncbi.nlm.nih.gov/substance/403459920
CONTENTS