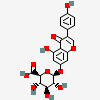
Genistein 7-O-glucuronide
PubChem CID
15940724
Molecular Formula
Synonyms
- 38482-81-4
- Genistein 7-O-glucuronide
- Genistein 7-|A-D-Glucuronide
- GENISTEIN 7-BETA-D-GLUCURONIDE
- genistein-7-o-glucuronide
Molecular Weight
446.4 g/mol
Computed by PubChem 2.2 (PubChem release 2024.11.20)
Dates
- Create:2007-02-21
- Modify:2025-01-18
Description
Genistein 7-O-glucuronide is an isoflavonoid and an acrovestone.
Genistein 7-O-glucuronide has been reported in Streptomyces lanatus with data available.
Chemical Structure Depiction
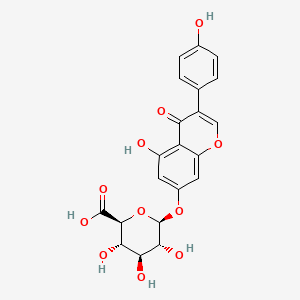
(2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-[5-hydroxy-3-(4-hydroxyphenyl)-4-oxochromen-7-yl]oxyoxane-2-carboxylic acid
Computed by Lexichem TK 2.7.0 (PubChem release 2024.11.20)
InChI=1S/C21H18O11/c22-9-3-1-8(2-4-9)11-7-30-13-6-10(5-12(23)14(13)15(11)24)31-21-18(27)16(25)17(26)19(32-21)20(28)29/h1-7,16-19,21-23,25-27H,(H,28,29)/t16-,17-,18+,19-,21+/m0/s1
Computed by InChI 1.07.0 (PubChem release 2024.11.20)
JIVINIISUDEORF-ZFORQUDYSA-N
Computed by InChI 1.07.0 (PubChem release 2024.11.20)
C1=CC(=CC=C1C2=COC3=CC(=CC(=C3C2=O)O)O[C@H]4[C@@H]([C@H]([C@@H]([C@H](O4)C(=O)O)O)O)O)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C21H18O11
Computed by PubChem 2.2 (PubChem release 2024.11.20)
38482-81-4
- 38482-81-4
- Genistein 7-O-glucuronide
- Genistein 7-|A-D-Glucuronide
- GENISTEIN 7-BETA-D-GLUCURONIDE
- genistein-7-o-glucuronide
- (2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-[5-hydroxy-3-(4-hydroxyphenyl)-4-oxochromen-7-yl]oxyoxane-2-carboxylic acid
- (2S,3S,4S,5R,6S)-3,4,5-Trihydroxy-6-((5-hydroxy-3-(4-hydroxyphenyl)-4-oxo-4H-chromen-7-yl)oxy)tetrahydro-2H-pyran-2-carboxylic acid
- Genistein 7-?-D-Glucuronide
- MEGxm0_000509
- CHEMBL3527014
- SCHEMBL13280116
- ACon0_001348
- ACon1_000168
- DTXSID60579964
- CHEBI:176131
- AKOS027447852
- 5-Hydroxy-3-(4-hydroxyphenyl)-4-oxo-4H-1-benzopyran-7-yl beta-D-glucopyranosiduronic acid
- NCGC00180816-01
- HY-137967
- CS-0143376
- BRD-K23022982-001-01-2
- Q27460582
- (2S,3S,4S,5R,6S)-3,4,5-Trihydroxy-6-((5-hydroxy-3-(4-hydroxyphenyl)-4-oxo-4H-chromen-7-yl)oxy)tetrahydro-2H-pyran-2-carboxylicacid
- (2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-{[5-hydroxy-3-(4-hydroxyphenyl)-4-oxo-4H-chromen-7-yl]oxy}oxane-2-carboxylic acid
- G7G
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
446.4 g/mol
Reference
Computed by PubChem 2.2 (PubChem release 2024.11.20)
Property Name
XLogP3-AA
Property Value
1.1
Reference
Computed by XLogP3 3.0 (PubChem release 2024.11.20)
Property Name
Hydrogen Bond Donor Count
Property Value
6
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2024.11.20)
Property Name
Hydrogen Bond Acceptor Count
Property Value
11
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2024.11.20)
Property Name
Rotatable Bond Count
Property Value
4
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2024.11.20)
Property Name
Exact Mass
Property Value
446.08491139 Da
Reference
Computed by PubChem 2.2 (PubChem release 2024.11.20)
Property Name
Monoisotopic Mass
Property Value
446.08491139 Da
Reference
Computed by PubChem 2.2 (PubChem release 2024.11.20)
Property Name
Topological Polar Surface Area
Property Value
183 Ų
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2024.11.20)
Property Name
Heavy Atom Count
Property Value
32
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
746
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2024.11.20)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
5
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2021.10.14)
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Parent, Connectivity Count
Similar Compounds (2D)
Similar Conformers (3D)
Same Count
Protein Structures Count
- Kidney
- Liver
- Cytoplasm
- Extracellular
The LOTUS Initiative for Open Natural Products Research: frozen dataset union wikidata (with metadata) | DOI:10.5281/zenodo.5794106
- ChEBIGenistein 7-O-glucuronidehttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:176131
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/Genistein 7-O-glucuronidehttps://www.wikidata.org/wiki/Q27460582LOTUS Treehttps://lotus.naturalproducts.net/
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.html
- EPA DSSTox5-Hydroxy-3-(4-hydroxyphenyl)-4-oxo-4H-1-benzopyran-7-yl beta-D-glucopyranosiduronic acidhttps://comptox.epa.gov/dashboard/DTXSID60579964CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingGenistein 7-O-glucuronidehttp://www.hmdb.ca/metabolites/HMDB0041739
- Japan Chemical Substance Dictionary (Nikkaji)
- Metabolomics WorkbenchGenistein 7-O-glucuronidehttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=49516
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Wikidatagenistein-7-O-glucuronidehttps://www.wikidata.org/wiki/Q27460582
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
CONTENTS