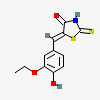
(5Z)-5-(3-ethoxy-4-hydroxybenzylidene)-2-sulfanyl-1,3-thiazol-4(5H)-one
PubChem CID
1389487
Molecular Formula
Synonyms
- (5Z)-5-(3-ethoxy-4-hydroxybenzylidene)-2-sulfanyl-1,3-thiazol-4(5H)-one
- (5Z)-5-[(3-ethoxy-4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one
- 6322-57-2
- 1287651-83-5
- (5~{Z})-5-[(3-ethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one
Molecular Weight
281.4 g/mol
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Dates
- Create:2005-07-11
- Modify:2025-01-11
Chemical Structure Depiction
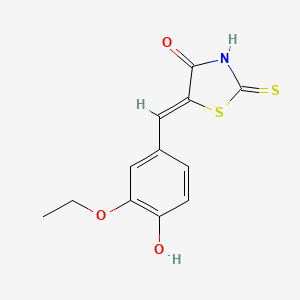
(5Z)-5-[(3-ethoxy-4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one
Computed by Lexichem TK 2.7.0 (PubChem release 2021.05.07)
InChI=1S/C12H11NO3S2/c1-2-16-9-5-7(3-4-8(9)14)6-10-11(15)13-12(17)18-10/h3-6,14H,2H2,1H3,(H,13,15,17)/b10-6-
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
BLHXTUCOFJOMCR-POHAHGRESA-N
Computed by InChI 1.0.6 (PubChem release 2021.05.07)
CCOC1=C(C=CC(=C1)/C=C\2/C(=O)NC(=S)S2)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C12H11NO3S2
Computed by PubChem 2.1 (PubChem release 2021.05.07)
6322-57-2
- (5Z)-5-(3-ethoxy-4-hydroxybenzylidene)-2-sulfanyl-1,3-thiazol-4(5H)-one
- (5Z)-5-[(3-ethoxy-4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one
- 6322-57-2
- 1287651-83-5
- (5~{Z})-5-[(3-ethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one
- NSC32114
- SCHEMBL8165608
- CHEMBL5188976
- BBL017093
- NSC-32114
- STK440671
- STK795739
- AKOS000607916
- AKOS003327744
- VS-05844
- SR-01000361532
- SR-01000361532-1
- (Z)-5-(3-ethoxy-4-hydroxybenzylidene)-2-thioxothiazolidin-4-one
- (5Z)-5-(3-ethoxy-4-hydroxybenzylidene)-2-thioxo-1,3-thiazolidin-4-one
- (5Z)-5-[(3-ethoxy-4-hydroxyphenyl)methylidene]-2-sulfanyl-4,5-dihydro-1,3-thiazol-4-one
- 4VZ
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
281.4 g/mol
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
XLogP3-AA
Property Value
2.8
Reference
Computed by XLogP3 3.0 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Donor Count
Property Value
2
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Hydrogen Bond Acceptor Count
Property Value
5
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Rotatable Bond Count
Property Value
3
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Exact Mass
Property Value
281.01803556 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Monoisotopic Mass
Property Value
281.01803556 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Topological Polar Surface Area
Property Value
116 Ų
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Heavy Atom Count
Property Value
18
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
384
Reference
Computed by Cactvs 3.4.8.18 (PubChem release 2021.05.07)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2011.04.04)
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Parent, Connectivity Count
Similar Compounds (2D)
Similar Conformers (3D)
Same Count
Protein Structures Count
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=BLHXTUCOFJOMCR-POHAHGRESA-N
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- DTP/NCILICENSEUnless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source.https://www.cancer.gov/policies/copyright-reuse
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 403797838https://pubchem.ncbi.nlm.nih.gov/substance/403797838
CONTENTS