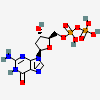
2'-Deoxyguanosine-5'-diphosphate
- dGDP
- 2'-deoxyguanosine-5'-diphosphate
- 3493-09-2
- CHEBI:28862
- 2'-Deoxyguanosine 5'-diphosphate
- Create:2019-01-15
- Modify:2025-01-18

- dGDP
- 2'-deoxyguanosine-5'-diphosphate
- 3493-09-2
- CHEBI:28862
- 2'-Deoxyguanosine 5'-diphosphate
- Deoxyguanosine 5'-diphosphate
- deoxyguanosine diphosphate
- CHEMBL1232205
- [(2R,3S,5R)-5-(2-amino-6-oxo-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methyl phosphono hydrogen phosphate
- 2'-deoxyguanosine 5'-(trihydrogen diphosphate)
- 2'-Deoxy-GDP
- deoxyguanosine-diphosphate
- 5'-dGDP
- [({[(2R,3S,5R)-5-(2-amino-6-oxo-6,9-dihydro-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]phosphonic acid
- SCHEMBL157937
- DTXSID601027110
- [[5-(2-amino-6-oxo-3H-purin-9-yl)-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxyphosphonic acid
- BDBM50035297
- DB03491
- PD059735
- NS00015101
- C00361
- Q27103932
- 2invertedexclamationmarka-Deoxyguanosine5invertedexclamationmarka-(trihydrogendiphosphate)
- 2 inverted exclamation marka-Deoxyguanosine 5 inverted exclamation marka-(trihydrogen diphosphate)
187.7 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with Agilent tune mix (Agilent)]
174.28 Ų [M-H2O-H]- [CCS Type: DT; Method: single field calibrated with Agilent tune mix (Agilent)]
188.5 Ų [M+H]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
185.6 Ų [M+Na-2H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
185.2 Ų [M+Na]+ [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
180.8 Ų [M-H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
177.5 Ų [M+Na-2H]- [CCS Type: DT; Method: single field calibrated with ESI Low Concentration Tuning Mix (Agilent)]
177.4 Ų [M-2H+Na]-
185.7 Ų [M-2H+Na]-
180.8 Ų [M-H]-
185.1 Ų [M+Na]+
188.5 Ų [M+H]+
78.9603 50.45
426.0216 45.59
158.9266 38.71
152.056549 100
81.034004 18.64
87.092079 5.05
113.107422 4.94
281.152191 4.82
78.9603 999
426.0216 903
158.9266 767
78.9603 50.45
426.0216 45.59
158.9266 38.71
152 100
151 20.51
81 6.80
426 100
425 19.52
427 14.33
- Cytoplasm
- Mitochondria
- Nucleus
- Adenine phosphoribosyltransferase deficiency (APRT)
- Adenosine Deaminase Deficiency
- Adenylosuccinate Lyase Deficiency
- AICA-Ribosiduria
- Azathioprine Action Pathway
- Gout or Kelley-Seegmiller Syndrome
- Lesch-Nyhan Syndrome (LNS)
- Mercaptopurine Action Pathway
- Mitochondrial DNA depletion syndrome
- Molybdenum Cofactor Deficiency
- Total 17 pathways, visit the HMDB page for details
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=CIKGWCTVFSRMJU-KVQBGUIXSA-N
- CAS Common ChemistryLICENSEThe data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc/4.0/
- EPA DSSTox[(2R,3S,5R)-5-(2-Amino-6-oxo-1H-purin-9-yl)-3-hydroxyoxolan-2-yl]methyl phosphono hydrogen phosphatehttps://comptox.epa.gov/dashboard/DTXSID601027110CompTox Chemicals Dashboard Chemical Listshttps://comptox.epa.gov/dashboard/chemical-lists/
- Human Metabolome Database (HMDB)LICENSEHMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications.http://www.hmdb.ca/citingHMDB0000960_msms_439183https://hmdb.ca/metabolites/HMDB0000960#spectra
- CCSbaseCCSbase Classificationhttps://ccsbase.net/
- NORMAN Suspect List ExchangeLICENSEData: CC-BY 4.0; Code (hosted by ECI, LCSB): Artistic-2.0https://creativecommons.org/licenses/by/4.0/Dgdp | 2'-deoxy-gdpNORMAN Suspect List Exchange Classificationhttps://www.norman-network.com/nds/SLE/
- ChEBI
- E. coli Metabolome Database (ECMDB)
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/2'-Deoxyguanosine-5'-diphosphatehttps://www.wikidata.org/wiki/Q27103932LOTUS Treehttps://lotus.naturalproducts.net/
- Yeast Metabolome Database (YMDB)
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.htmlChEMBL Protein Target Treehttps://www.ebi.ac.uk/chembl/g/#browse/targets
- DrugBankLICENSECreative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode)https://www.drugbank.ca/legal/terms_of_use2'-Deoxyguanosine-5'-Diphosphatehttps://www.drugbank.ca/drugs/DB03491
- FooDBLICENSEFooDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (FooDB) and the original publication.https://foodb.ca/about
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.htmlCompounds with biological roleshttp://www.genome.jp/kegg-bin/get_htext?br08001.keg
- MassBank Europe2'-Deoxyguanosine-5'-diphosphatehttps://massbank.eu/MassBank/Result.jsp?inchikey=CIKGWCTVFSRMJU-KVQBGUIXSA-N
- MassBank of North America (MoNA)LICENSEThe content of the MoNA database is licensed under CC BY 4.0.https://mona.fiehnlab.ucdavis.edu/documentation/license
- Metabolomics Workbench
- Protein Data Bank in Europe (PDBe)
- RCSB Protein Data Bank (RCSB PDB)LICENSEData files contained in the PDB archive (ftp://ftp.wwpdb.org) are free of all copyright restrictions and made fully and freely available for both non-commercial and commercial use. Users of the data should attribute the original authors of that structural data.https://www.rcsb.org/pages/policies
- Springer Nature
- Wikidata
- WikipediaDeoxyguanosine diphosphatehttps://en.wikipedia.org/wiki/Deoxyguanosine_diphosphate
- PubChem
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 388558815https://pubchem.ncbi.nlm.nih.gov/substance/388558815