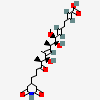
13-epi-dorrigocin A
PubChem CID
44543739
Molecular Formula
Synonyms
- 13-epi-Dorrigocin A
- (2E,6E,8S,9S,10R,11E,13S,14S)-18-(2,6-dioxopiperidin-4-yl)-9,13-dihydroxy-8-methoxy-10,12,14-trimethyl-15-oxooctadeca-2,6,11-trienoic acid
- CHEMBL474522
- CHEBI:219677
- NCGC00488794-01
Molecular Weight
507.6 g/mol
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Dates
- Create:2010-01-19
- Modify:2025-01-10
Description
13-epi-dorrigocin A is a lipid.
13-epi-Dorrigocin A has been reported in Streptomyces platensis with data available.
Chemical Structure Depiction

Conformer generation is disallowed since too flexible
(2E,6E,8S,9S,10R,11E,13S,14S)-18-(2,6-dioxopiperidin-4-yl)-9,13-dihydroxy-8-methoxy-10,12,14-trimethyl-15-oxooctadeca-2,6,11-trienoic acid
Computed by LexiChem 2.6.6 (PubChem release 2019.06.18)
InChI=1S/C27H41NO8/c1-17(14-18(2)27(35)22(36-4)12-7-5-6-8-13-25(32)33)26(34)19(3)21(29)11-9-10-20-15-23(30)28-24(31)16-20/h7-8,12-14,18-20,22,26-27,34-35H,5-6,9-11,15-16H2,1-4H3,(H,32,33)(H,28,30,31)/b12-7+,13-8+,17-14+/t18-,19-,22+,26-,27+/m1/s1
Computed by InChI 1.0.5 (PubChem release 2019.06.18)
HJCZOTBHYAPUHT-FFMZNCFZSA-N
Computed by InChI 1.0.5 (PubChem release 2019.06.18)
C[C@H](/C=C(\C)/[C@H]([C@H](C)C(=O)CCCC1CC(=O)NC(=O)C1)O)[C@@H]([C@H](/C=C/CC/C=C/C(=O)O)OC)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C27H41NO8
Computed by PubChem 2.1 (PubChem release 2019.06.18)
- 13-epi-Dorrigocin A
- (2E,6E,8S,9S,10R,11E,13S,14S)-18-(2,6-dioxopiperidin-4-yl)-9,13-dihydroxy-8-methoxy-10,12,14-trimethyl-15-oxooctadeca-2,6,11-trienoic acid
- CHEMBL474522
- CHEBI:219677
- NCGC00488794-01
- (2E,6E,8S,9S,10R,13S,14S)-9,13-Dihydroxy-18-(6-hydroxy-2-oxo-2,3,4,5-tetrahydropyridin-4-yl)-8-methoxy-10,12,14-trimethyl-15-oxooctadeca-2,6,11-trienoate
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
507.6 g/mol
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
XLogP3-AA
Property Value
1.7
Reference
Computed by XLogP3 3.0 (PubChem release 2019.06.18)
Property Name
Hydrogen Bond Donor Count
Property Value
4
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Hydrogen Bond Acceptor Count
Property Value
8
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Rotatable Bond Count
Property Value
16
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Exact Mass
Property Value
507.28321727 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Monoisotopic Mass
Property Value
507.28321727 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Topological Polar Surface Area
Property Value
150 Ų
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Heavy Atom Count
Property Value
36
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
831
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
5
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
3
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2019.01.04)
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Parent, Connectivity Count
Similar Compounds (2D)
Similar Conformers (3D)
Same Count
The LOTUS Initiative for Open Natural Products Research: frozen dataset union wikidata (with metadata) | DOI:10.5281/zenodo.5794106
- ChEBI13-epi-dorrigocin Ahttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:219677
- LOTUS - the natural products occurrence databaseLICENSEThe code for LOTUS is released under the GNU General Public License v3.0.https://lotus.nprod.net/13-epi-Dorrigocin Ahttps://www.wikidata.org/wiki/Q77568134LOTUS Treehttps://lotus.naturalproducts.net/
- ChEMBLLICENSEAccess to the web interface of ChEMBL is made under the EBI's Terms of Use (http://www.ebi.ac.uk/Information/termsofuse.html). The ChEMBL data is made available on a Creative Commons Attribution-Share Alike 3.0 Unported License (http://creativecommons.org/licenses/by-sa/3.0/).http://www.ebi.ac.uk/Information/termsofuse.html
- Japan Chemical Substance Dictionary (Nikkaji)
- Metabolomics Workbench13-epi-dorrigocin Ahttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=103351
- Nature Chemical Biology
- Springer Nature
- Wikidata13-epi-dorrigocin Ahttps://www.wikidata.org/wiki/Q77568134
- PubChem
- The Natural Products AtlasLICENSEThe Natural Products Atlas is licensed under a Creative Commons Attribution 4.0 International License.https://www.npatlas.org/termsThe Natural Products Atlas Classificationhttps://www.npatlas.org/
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
CONTENTS