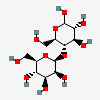
4-O-beta-D-mannopyranosyl-D-glucopyranose
PubChem CID
10450027
Molecular Formula
Synonyms
- 4-O-beta-D-mannopyranosyl-D-glucopyranose
- 29276-55-9
- Manbeta1->4Glc
- mannosylglucose
- beta-D-Manp-(1->4)-D-Glcp
Molecular Weight
342.30 g/mol
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Dates
- Create:2006-10-25
- Modify:2025-01-18
Chemical Structure Depiction
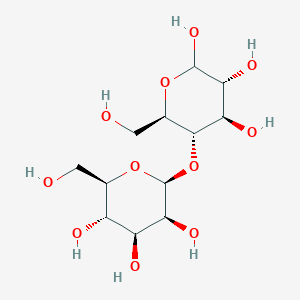
SVG Image

IUPAC Condensed
Man(b1-4)Glc
LINUCS
[][D-Glcp]{[(4+1)][b-D-Manp]{}}
IUPAC
beta-D-manno-hexopyranosyl-(1->4)-D-gluco-hexopyranose
(2R,3S,4S,5S,6S)-2-(hydroxymethyl)-6-[(2R,3S,4R,5R)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxyoxane-3,4,5-triol
Computed by LexiChem 2.6.6 (PubChem release 2019.06.18)
InChI=1S/C12H22O11/c13-1-3-5(15)6(16)9(19)12(22-3)23-10-4(2-14)21-11(20)8(18)7(10)17/h3-20H,1-2H2/t3-,4-,5-,6+,7-,8-,9+,10-,11?,12+/m1/s1
Computed by InChI 1.0.5 (PubChem release 2019.06.18)
GUBGYTABKSRVRQ-OKIQBEFVSA-N
Computed by InChI 1.0.5 (PubChem release 2019.06.18)
C([C@@H]1[C@H]([C@@H]([C@@H]([C@@H](O1)O[C@@H]2[C@H](OC([C@@H]([C@H]2O)O)O)CO)O)O)O)O
Computed by OEChem 2.3.0 (PubChem release 2024.12.12)
C12H22O11
Computed by PubChem 2.1 (PubChem release 2019.06.18)
- 4-O-beta-D-mannopyranosyl-D-glucopyranose
- 29276-55-9
- Manbeta1->4Glc
- mannosylglucose
- beta-D-Manp-(1->4)-D-Glcp
- (Glc)1 (Man)1
- beta-D-mannosyl-(1->4)-D-glucose
- beta-D-Man-(1->4)-D-Glc
- 4-O-BETA-D-MANNOPYRANOSYL-D-GLUCOPYRANOSIDE
- CHEBI:64351
- beta-D-mannopyranosyl-(1->4)-D-glucopyranose
- (2R,3S,4S,5S,6S)-2-(hydroxymethyl)-6-[(2R,3S,4R,5R)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxyoxane-3,4,5-triol
- 4-O-(b-D-Mannopyranosyl)-D-glucose
- 28072-80-2
- 4-O-B-D-MANNOPYRANOSYL-D-GLUCOPYRANOSIDE
- 4-O-(beta-D-Mannopyranosyl)-D-glucose
- Man1-b-4-Glc
- 4-O-?-D-Mannopyranosyl-D-glucopyranoside
- C20236
- Q27133233
- (3R,4R,5S,6R)-6-(Hydroxymethyl)-5-(((2S,3S,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)tetrahydro-2H-pyran-2,3,4-triol
Property Name
Property Value
Reference
Property Name
Molecular Weight
Property Value
342.30 g/mol
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
XLogP3-AA
Property Value
-4.7
Reference
Computed by XLogP3 3.0 (PubChem release 2019.06.18)
Property Name
Hydrogen Bond Donor Count
Property Value
8
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Hydrogen Bond Acceptor Count
Property Value
11
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Rotatable Bond Count
Property Value
4
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Exact Mass
Property Value
342.11621151 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Monoisotopic Mass
Property Value
342.11621151 Da
Reference
Computed by PubChem 2.1 (PubChem release 2021.05.07)
Property Name
Topological Polar Surface Area
Property Value
190 Ų
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Heavy Atom Count
Property Value
23
Reference
Computed by PubChem
Property Name
Formal Charge
Property Value
0
Reference
Computed by PubChem
Property Name
Complexity
Property Value
382
Reference
Computed by Cactvs 3.4.6.11 (PubChem release 2019.06.18)
Property Name
Isotope Atom Count
Property Value
0
Reference
Computed by PubChem
Property Name
Defined Atom Stereocenter Count
Property Value
9
Reference
Computed by PubChem
Property Name
Undefined Atom Stereocenter Count
Property Value
1
Reference
Computed by PubChem
Property Name
Defined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Undefined Bond Stereocenter Count
Property Value
0
Reference
Computed by PubChem
Property Name
Covalently-Bonded Unit Count
Property Value
1
Reference
Computed by PubChem
Property Name
Compound Is Canonicalized
Property Value
Yes
Reference
Computed by PubChem (release 2019.01.04)
Follow these links to do a live 2D search or do a live 3D search for this compound, sorted by annotation score. This section is deprecated (see here for details), but these live search links provide equivalent functionality to the table that was previously shown here.
Same Connectivity Count
Same Isotope Count
Same Parent, Connectivity Count
Same Parent, Isotope Count
Similar Compounds (2D)
Similar Conformers (3D)
Same Count
PubMed Count
Patents are available for this chemical structure:
https://patentscope.wipo.int/search/en/result.jsf?inchikey=GUBGYTABKSRVRQ-OKIQBEFVSA-N
- ChEBIBeta-D-Manp-(1->4)-D-Glcphttps://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:64351
- Japan Chemical Substance Dictionary (Nikkaji)
- KEGGLICENSEAcademic users may freely use the KEGG website. Non-academic use of KEGG generally requires a commercial licensehttps://www.kegg.jp/kegg/legal.html
- Metabolomics Workbenchbeta-D-Manp-(1->4)-D-Glcphttps://www.metabolomicsworkbench.org/data/StructureData.php?RegNo=63340
- Rhea - Annotated Reactions DatabaseLICENSERhea has chosen to apply the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). This means that you are free to copy, distribute, display and make commercial use of the database in all legislations, provided you credit (cite) Rhea.https://www.rhea-db.org/help/license-disclaimer
- Thieme ChemistryLICENSEThe Thieme Chemistry contribution within PubChem is provided under a CC-BY-NC-ND 4.0 license, unless otherwise stated.https://creativecommons.org/licenses/by-nc-nd/4.0/
- Wikidatabeta-D-Manp-(1->4)-D-Glcphttps://www.wikidata.org/wiki/Q27133233
- PubChem
- Glycan Naming and Subsumption Ontology (GNOme)GNOme
- MolGenieMolGenie Organic Chemistry Ontologyhttps://github.com/MolGenie/ontology/
- PATENTSCOPE (WIPO)SID 504991160https://pubchem.ncbi.nlm.nih.gov/substance/504991160
CONTENTS